1IPA
 
 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
1RRB
 
 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1GE6
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE5
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE7
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2CYZ
 
 | | photo-activation state of Fe-type NHase in anaerobic condition | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | photo-activation state of Fe-type NHase in anaerobic condition
To be Published
|
|
2CZ1
 
 | | photo-activation state of Fe-type NHase with n-BA in anaerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | photo-activation state of Fe-type NHase with n-BA in anaerobic condition
To be Published
|
|
2CZ0
 
 | | photo-activation state of Fe-type NHase in aerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | photo-activation state of Fe-type NHase in aerobic condition
To be Published
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
2ZIY
 
 | | Crystal structure of squid rhodopsin | | Descriptor: | PALMITIC ACID, RETINAL, Rhodopsin | | Authors: | Miyano, M, Shimamura, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region
J.Biol.Chem., 283, 2008
|
|
2ZTI
 
 | |
1ODJ
 
 | |
1ODI
 
 | |
1ODL
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, GLYCEROL, PURINE NUCLEOSIDE PHOSPHORYLASE, ... | | Authors: | Tahirov, T.H, Inagaki, E, Miyano, M. | | Deposit date: | 2003-02-19 | | Release date: | 2003-02-27 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase from Thermus Thermophilus
J.Mol.Biol., 337, 2004
|
|
1ODK
 
 | |
1V1A
 
 | |
1V1B
 
 | |
1V1S
 
 | |
1V19
 
 | | 2-KETO-3-DEOXYGLUCONATE KINASE FROM THERMUS THERMOPHILUS | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-KETO-3-DEOXYGLUCONATE KINASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Thermus Thermophilus 2-Keto-3-Deoxygluconate Kinase: Evidence for Recognition of an Open Chain Substrate
J.Mol.Biol., 340, 2004
|
|
1V7R
 
 | |
1WR8
 
 | |
1WP4
 
 | |
1DLF
 
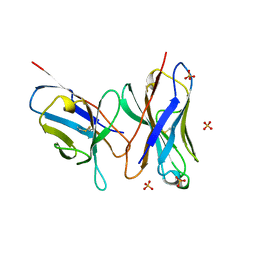 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
