1D1H
 
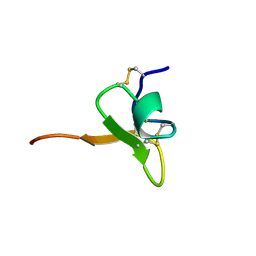 | | SOLUTION STRUCTURE OF HANATOXIN 1 | | Descriptor: | HANATOXIN TYPE 1 | | Authors: | Takahashi, H, Kim, J.I, Sato, K, Swartz, K.J, Shimada, I. | | Deposit date: | 1999-09-16 | | Release date: | 2000-09-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: common surface features of gating modifier toxins.
J.Mol.Biol., 297, 2000
|
|
8DXQ
 
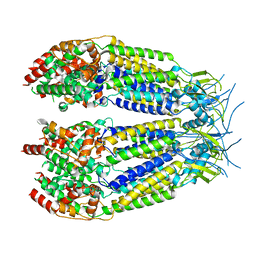 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 4 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXP
 
 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 3 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXO
 
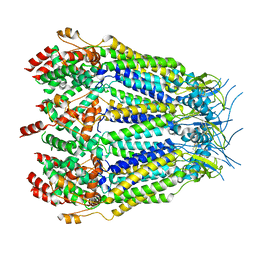 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 2 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXN
 
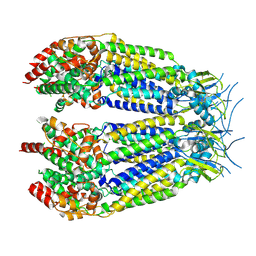 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 1 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXR
 
 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 5 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OI7
 
 | | The Crystal Structure of Succinyl-CoA synthetase alpha subunit from Thermus Thermophilus | | Descriptor: | SUCCINYL-COA SYNTHETASE ALPHA CHAIN | | Authors: | Takahashi, H, Tokunaga, Y, Kuroishi, C, Babayeva, N, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The Crystal Structure of Succinyl-Coa Synthetase from Thermus Thermophilus
To be Published
|
|
1UZ5
 
 | |
1US4
 
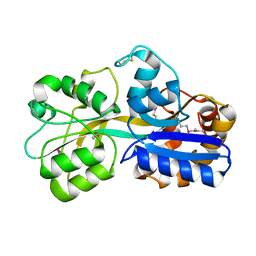 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E, Takahashi, H. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1USF
 
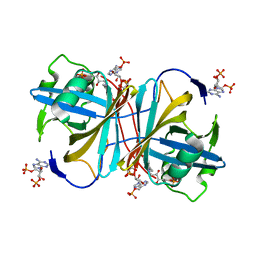 | |
1DLF
 
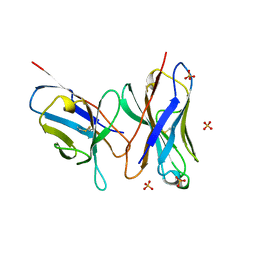 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
7T3R
 
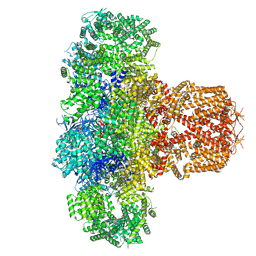 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active C state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3U
 
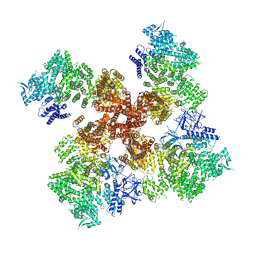 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3P
 
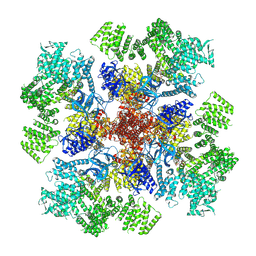 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active A state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3T
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3Q
 
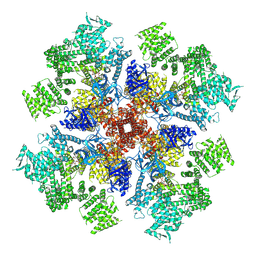 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active B state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
3OTJ
 
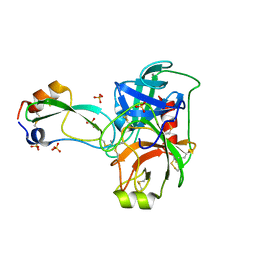 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3FHP
 
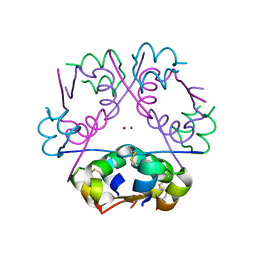 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2DLF
 
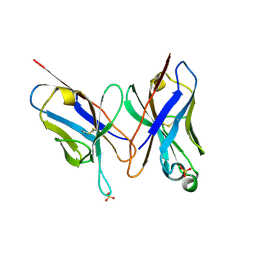 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2RSG
 
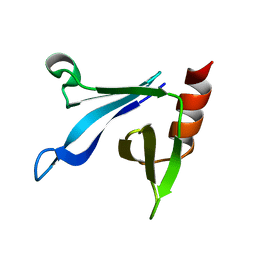 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
1USC
 
 | |
1WZ1
 
 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | Descriptor: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | Authors: | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | Deposit date: | 2005-02-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
1KOZ
 
 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | Descriptor: | Voltage-dependent Channel Inhibitor | | Authors: | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | Deposit date: | 2001-12-25 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
