4WAS
 
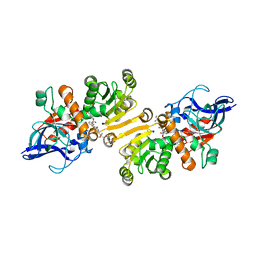 | | STRUCTURE OF THE ETR1P/NADP/CROTONYL-COA COMPLEX | | Descriptor: | CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Capitani, G, Erb, T.J. | | Deposit date: | 2014-08-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
4WJ7
 
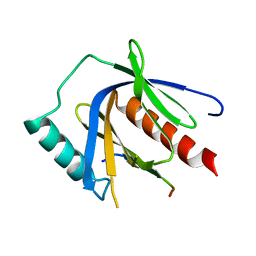 | | CCM2 PTB domain in complex with KRIT1 NPxY/F3 | | Descriptor: | KRIT1 NPxY/F3, Malcavernin | | Authors: | Fisher, O.S, Liu, W, Zhang, R, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structural Basis for the Disruption of the Cerebral Cavernous Malformations 2 (CCM2) Interaction with Krev Interaction Trapped 1 (KRIT1) by Disease-associated Mutations.
J.Biol.Chem., 290, 2015
|
|
1A62
 
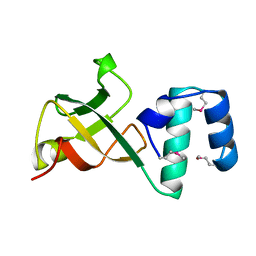 | | CRYSTAL STRUCTURE OF THE RNA-BINDING DOMAIN OF THE TRANSCRIPTIONAL TERMINATOR PROTEIN RHO | | Descriptor: | RHO | | Authors: | Allison, T.J, Wood, T.C, Briercheck, D.M, Rastinejad, F, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the RNA-binding domain from transcription termination factor rho.
Nat.Struct.Biol., 5, 1998
|
|
1A63
 
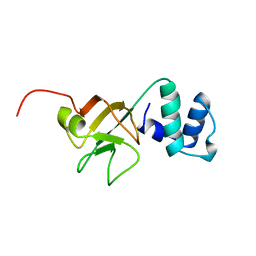 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
1AUP
 
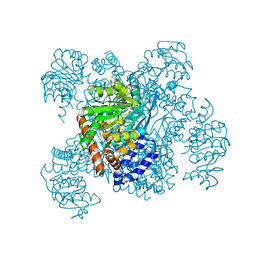 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1BGV
 
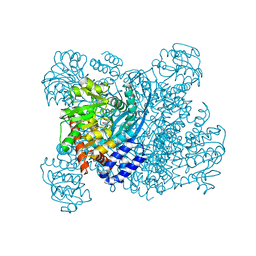 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
1A6T
 
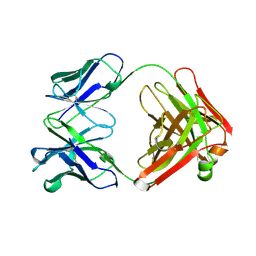 | |
1AN5
 
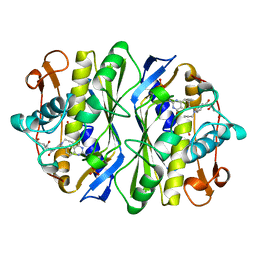 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1AF8
 
 | | ACTINORHODIN POLYKETIDE SYNTHASE ACYL CARRIER PROTEIN FROM STREPTOMYCES COELICOLOR A3(2), NMR, 24 STRUCTURES | | Descriptor: | ACTINORHODIN POLYKETIDE SYNTHASE ACYL CARRIER PROTEIN | | Authors: | Crump, M.P, Crosby, J, Dempsey, C.E, Parkinson, J.A, Murray, M, Hopwood, D.A, Simpson, T.J. | | Deposit date: | 1997-03-23 | | Release date: | 1997-09-26 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2).
Biochemistry, 36, 1997
|
|
1B3D
 
 | | STROMELYSIN-1 | | Descriptor: | CALCIUM ION, N-[[2-METHYL-4-HYDROXYCARBAMOYL]BUT-4-YL-N]-BENZYL-P-[PHENYL]-P-[METHYL]PHOSPHINAMID, STROMELYSIN-1, ... | | Authors: | Chen, L, Rydel, T.J, Dunaway, C.M, Pikul, S, Dunham, K.M, Gu, F, Barnett, B.L. | | Deposit date: | 1998-12-09 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the stromelysin catalytic domain at 2.0 A resolution: inhibitor-induced conformational changes.
J.Mol.Biol., 293, 1999
|
|
1B0Q
 
 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
1BVU
 
 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
3L6V
 
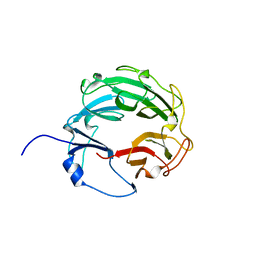 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
1K89
 
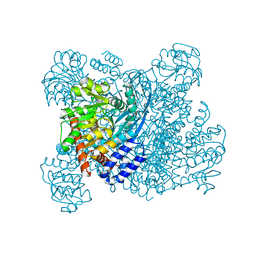 | | K89L MUTANT OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Stillman, T.J, Migueis, A.M.B, Wang, X.G, Baker, P.J, Britton, K.L, Engel, P.C, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the mechanism of domain closure and substrate specificity of glutamate dehydrogenase from Clostridium symbiosum.
J.Mol.Biol., 285, 1999
|
|
6GKI
 
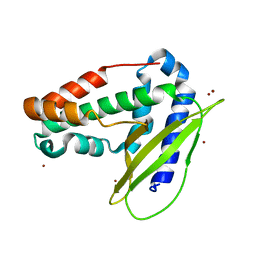 | | Structure of E coli MlaC in Variously Loaded States | | Descriptor: | BROMIDE ION, GLYCEROL, Probable phospholipid-binding protein MlaC | | Authors: | Knowles, T.J, Lovering, A.L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system.
Nat Microbiol, 4, 2019
|
|
3FFD
 
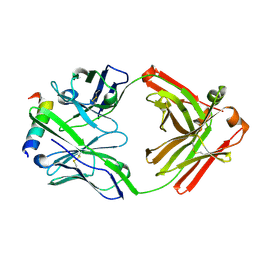 | | Structure of parathyroid hormone-related protein complexed to a neutralizing monoclonal antibody | | Descriptor: | Monoclonal antibody, heavy chain, Fab fragment, ... | | Authors: | Mckinstry, W.J, Polekhina, G, Diefenbach-Jagger, H, Ho, P.W.M, Sato, K, Onuma, E, Gillespie, M.T, Martin, T.J, Parker, M.W. | | Deposit date: | 2008-12-03 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor
J.Biol.Chem., 284, 2009
|
|
1BM3
 
 | | IMMUNOGLOBULIN OPG2 FAB-PEPTIDE COMPLEX | | Descriptor: | IMMUNOGLOBULIN OPG2 FAB, CONSTANT DOMAIN, VARIABLE DOMAIN | | Authors: | Kodandapani, R, Veerapandian, L, Ni, C.Z, Chiou, C.-K, Whital, R, Kunicki, T.J, Ely, K.R. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational change in an anti-integrin antibody: structure of OPG2 Fab bound to a beta 3 peptide.
Biochem.Biophys.Res.Commun., 251, 1998
|
|
7WUK
 
 | |
8AFV
 
 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | N-acetyl-gamma-glutamyl-phosphate reductase of Denitrovibrio acetiphilus
To Be Published
|
|
8AFU
 
 | |
8APR
 
 | |
8APQ
 
 | | CaMct - Mesaconyl-CoA C1:C4 CoA Transferase of Chloroflexus aurantiacus | | Descriptor: | (2E)-2-METHYLBUT-2-ENEDIOIC ACID, 2-methylfumaryl-CoA isomerase, COENZYME A, ... | | Authors: | Pfister, P, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-08-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis for a Cork-Up Mechanism of the Intra-Molecular Mesaconyl-CoA Transferase.
Biochemistry, 62, 2023
|
|
3FLR
 
 | |
