6VEJ
 
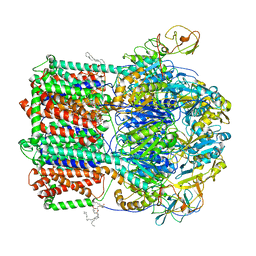 | | TriABC transporter from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(hexadecanoyloxy)propyl nonadecanoate, DODECYL-ALPHA-D-MALTOSIDE, PALMITIC ACID, ... | | Authors: | Sygusch, J, Fabre, L, Rouiller, I, Bhattacharyya, S. | | Deposit date: | 2020-01-02 | | Release date: | 2020-09-30 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A "Drug Sweeping" State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa.
Structure, 29, 2021
|
|
5VJD
 
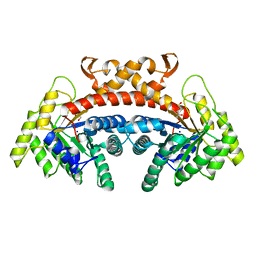 | | Class II fructose-1,6-bisphosphate aldolase of Escherichia coli with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase class 2, SODIUM ION, ... | | Authors: | Sygusch, J, Coincon, M. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
4XBH
 
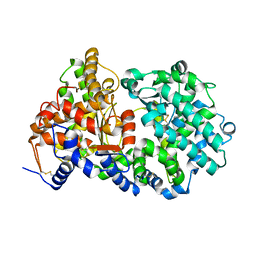 | | Soluble rabbit neprilysin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1ADO
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ALDOLASE, SULFATE ION | | Authors: | Blom, N.S, Sygusch, J. | | Deposit date: | 1996-12-02 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 4, 1997
|
|
5I97
 
 | |
5JBS
 
 | | Conformational changes during monomer-to-dimer transition of Brucella suis VirB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arya, T, Sharifahmadian, M, Sygusch, J, Baron, B. | | Deposit date: | 2016-04-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR analyses, X-ray crystallography and small-molecule probing reveal conformational shifts during monomer-to-dimer transition of Brucella suis VirB8
To Be Published
|
|
1L3A
 
 | |
5C17
 
 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0U
 
 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0T
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
1ZAI
 
 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from rabbit muscle | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAJ
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with mannitol-1,6-bisphosphate, a competitive inhibitor | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAH
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAL
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1FDJ
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT LIVER | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE, ... | | Authors: | Blom, N.S, White, A, Sygusch, J. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction intermediates of Rabbit liver D-fructose 1,6-bisphosphate Aldolase
To be Published
|
|
1RVG
 
 | |
1RV8
 
 | | Class II fructose-1,6-bisphosphate aldolase from Thermus aquaticus in complex with cobalt | | Descriptor: | COBALT (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Izard, T, Sygusch, J. | | Deposit date: | 2003-12-12 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induced Fit Movements and Metal Cofactor Selectivity of Class II Aldolases: STRUCTURE OF THERMUS AQUATICUS FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE.
J.Biol.Chem., 279, 2004
|
|
3LGE
 
 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
4KOO
 
 | | Crystal Structure of WHY1 from Arabidopsis thaliana | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOP
 
 | | Crystal Structure of WHY2 from Arabidopsis thaliana | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Single-stranded DNA-binding protein WHY2, mitochondrial | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOQ
 
 | | Crystal Structure of WHY3 from Arabidopsis thaliana | | Descriptor: | PHOSPHATE ION, Single-stranded DNA-binding protein WHY3, chloroplastic | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4A22
 
 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to N-(4-hydroxybutyl)- glycolohydroxamic acid bis- phosphate | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, ... | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli.
J. Biol. Chem., 286, 2011
|
|
4ZR5
 
 | | Soluble rabbit neprilysin in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2015-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8016 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
1DOS
 
 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | Descriptor: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | Authors: | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | Deposit date: | 1996-06-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
