1K0S
 
 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
2NP8
 
 | | Structural Basis for the Inhibition of Aurora A Kinase by a Novel Class of High Affinity Disubstituted Pyrimidine Inhibitors | | Descriptor: | N-{3-[(4-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}PYRIMIDIN-2-YL)AMINO]PHENYL}CYCLOPROPANECARBOXAMIDE, SULFATE ION, Serine/threonine-protein kinase 6 | | Authors: | Tari, L.W, Hoffman, I.D, Bensen, D.C, Hunter, M.J, Nix, J, Nelson, K.J, McRee, D.E, Swanson, R.V. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity disubstituted pyrimidine inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3HFB
 
 | |
3HF6
 
 | | Crystal structure of human tryptophan hydroxylase type 1 with bound LP-521834 and FE | | Descriptor: | 4-(4-amino-6-{[(1R)-1-naphthalen-2-ylethyl]amino}-1,3,5-triazin-2-yl)-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M.J. | | Deposit date: | 2009-05-11 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis
Curr Chem Genomics, 4, 2010
|
|
3HF8
 
 | | Crystal structure of human tryoptophan hydroxylase type 1 with bound LP-533401 and Fe | | Descriptor: | 4-{2-amino-6-[(1R)-2,2,2-trifluoro-1-(3'-fluorobiphenyl-4-yl)ethoxy]pyrimidin-4-yl}-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M.J. | | Deposit date: | 2009-05-11 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis.
Curr Chem Genomics, 4, 2010
|
|
3IPY
 
 | | X-Ray structure of Human Deoxycytidine Kinase in complex with an inhibitor | | Descriptor: | 4-(1-benzothiophen-2-yl)-6-[4-(2-oxo-2-pyrrolidin-1-ylethyl)piperazin-1-yl]pyrimidine, D-MALATE, Deoxycytidine kinase | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M, Hoffman, I, Stouch, T.R, Carson, K.G. | | Deposit date: | 2009-08-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Lead optimization and structure-based design of potent and bioavailable deoxycytidine kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IPX
 
 | | X-Ray structure of Human Deoxycytidine Kinase in complex with ADP and an inhibitor | | Descriptor: | 2'-deoxy-5-fluorocytidine, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, ... | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M, Hoffman, I, Stouch, T.R, Carson, K.G. | | Deposit date: | 2009-08-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead optimization and structure-based design of potent and bioavailable deoxycytidine kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1RQJ
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
1RTR
 
 | | Crystal Structure of S. Aureus Farnesyl Pyrophosphate Synthase | | Descriptor: | geranyltranstransferase | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Mol.Biol., 279, 2004
|
|
1T69
 
 | | Crystal Structure of human HDAC8 complexed with SAHA | | Descriptor: | Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ZINC ION | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1T67
 
 | | Crystal Structure of Human HDAC8 complexed with MS-344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1T64
 
 | | Crystal Structure of human HDAC8 complexed with Trichostatin A | | Descriptor: | CALCIUM ION, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1VKG
 
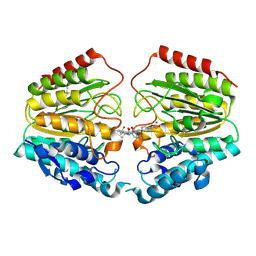 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1MP8
 
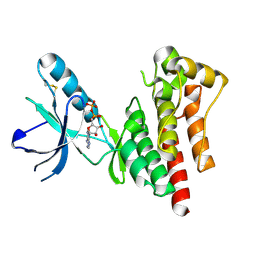 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
1MQ4
 
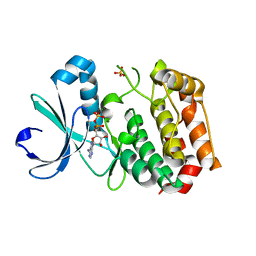 | | Crystal Structure of Aurora-A Protein Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA-RELATED KINASE 1, MAGNESIUM ION, ... | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-13 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Cancer-Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2002
|
|
1MQB
 
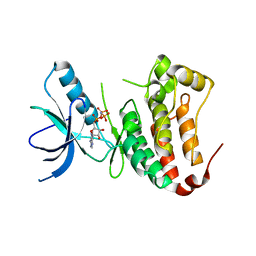 | | Crystal Structure of Ephrin A2 (ephA2) Receptor Protein Kinase | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N, Rogers, J, Sang, B.C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Cancer Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2003
|
|
1P3D
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P31
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P74
 
 | | CRYSTAL STRUCTURE OF SHIKIMATE DEHYDROGENASE (AROE) FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | Shikimate 5-dehydrogenase | | Authors: | Ye, S, von Delft, F, Brooun, A, Knuth, M.W, Swanson, R.V, McRee, D.E. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode
J.Bacteriol., 185, 2003
|
|
1P77
 
 | | CRYSTAL STRUCTURE OF SHIKIMATE DEHYDROGENASE (AROE) FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, ACETATE ION, Shikimate 5-dehydrogenase | | Authors: | Ye, S, von Delft, F, Brooun, A, Knuth, M.W, Swanson, R.V, McRee, D.E. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode
J.Bacteriol., 185, 2003
|
|
1QVB
 
 | | CRYSTAL STRUCTURE OF THE BETA-GLYCOSIDASE FROM THE HYPERTHERMOPHILE THERMOSPHAERA AGGREGANS | | Descriptor: | BETA-GLYCOSIDASE | | Authors: | Chi, Y.-I, Martinez-Cruz, L.A, Swanson, R.V, Robertson, D.E, Kim, S.-H. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability.
FEBS Lett., 445, 1999
|
|
1RQI
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Dimethylallyl S-Thiolodiphosphate | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
3L5X
 
 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
