1DRS
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE RGD-CONTAINING NEUROTOXIN HOMOLOGUE, DENDROASPIN | | Descriptor: | DENDROASPIN | | Authors: | Sutcliffe, M.J, Jaseja, M, Hyde, E.I, Lu, X, Williams, J.A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the RGD-containing neurotoxin homologue dendroaspin.
Nat.Struct.Biol., 1, 1994
|
|
2NBT
 
 | | NEURONAL BUNGAROTOXIN, NMR, 10 STRUCTURES | | Descriptor: | NEURONAL BUNGAROTOXIN | | Authors: | Oswald, R.E, Sutcliffe, M.J, Bamberger, M, Loring, R.H, Braswell, E, Dobson, C.M. | | Deposit date: | 1997-10-29 | | Release date: | 1998-03-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics.
Biochemistry, 31, 1992
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
1BBA
 
 | |
1EES
 
 | | SOLUTION STRUCTURE OF CDC42HS COMPLEXED WITH A PEPTIDE DERIVED FROM P-21 ACTIVATED KINASE, NMR, 20 STRUCTURES | | Descriptor: | GTP-BINDING PROTEIN, P21-ACTIVATED KINASE | | Authors: | Gizachew, D, Guo, W, Chohan, K.C, Sutcliffe, M.J, Oswald, R.E. | | Deposit date: | 2000-02-02 | | Release date: | 2000-03-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase.
Biochemistry, 39, 2000
|
|
1T9G
 
 | | Structure of the human MCAD:ETF complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, van Thiel, A, Basran, J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Extensive domain motion and electron transfer in the human electron transferring flavoprotein-medium chain Acyl-CoA dehydrogenase complex
J.Biol.Chem., 279, 2004
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
2IGG
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
2AH1
 
 | | Crystal structure of aromatic amine dehydrogenase (AADH) from Alcaligenes faecalis | | Descriptor: | Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGZ
 
 | | Crystal structure of the carbinolamine intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. F222 form | | Descriptor: | (1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL, Aromatic amine dehydrogenase, ZINC ION | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGX
 
 | | Crystal structure of the Schiff base intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. P212121 form | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGW
 
 | | Crystal structure of tryptamine-reduced aromatic amine dehydrogenase (AADH) from Alcaligenes faecalis in complex with tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AH0
 
 | | Crystal structure of the carbinolamine intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. Monoclinic form | | Descriptor: | (1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL, 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGL
 
 | | Crystal structure of the phenylhydrazine adduct of aromatic amine dehydrogenase from Alcaligenes faecalis | | Descriptor: | 1-PHENYLHYDRAZINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGY
 
 | | Crystal structure of the Schiff base intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. Monoclinic form | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
1O94
 
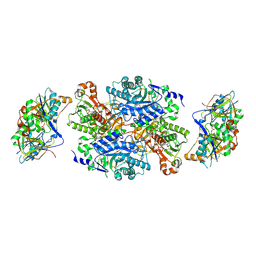 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O95
 
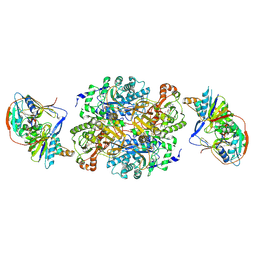 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O96
 
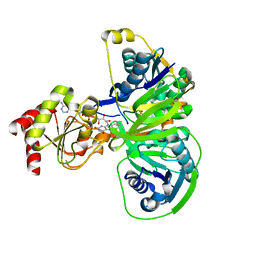 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O97
 
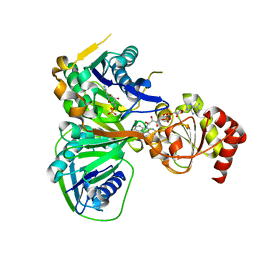 | | Structure of electron transferring flavoprotein from Methylophilus methylotrophus, recognition loop removed by limited proteolysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
2IGH
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
3GX9
 
 | | Structure of morphinone reductase N189A mutant in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Parallel Pathways and Free-Energy Landscapes for Enzymatic Hydride Transfer Probed by Hydrostatic Pressure
Chembiochem, 10, 2009
|
|
3KFT
 
 | | Crystal structure of Pentaerythritol Tetranitrate Reductase complex with 1,4,5,6-tetrahydro NADH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Pudney, C.R, Levy, C.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-10-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction.
J.Am.Chem.Soc., 131, 2009
|
|
2OK6
 
 | |
2OIZ
 
 | |
