5MUV
 
 | | Atomic structure fitted into a localized reconstruction of bacteriophage phi6 packaging hexamer P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
5MUW
 
 | | Atomic structure of P4 packaging enzyme fitted into a localized reconstruction of bacteriophage phi6 vertex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
5MUU
 
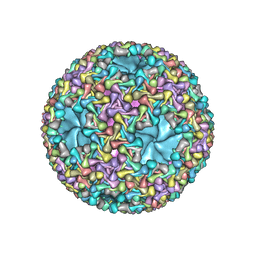 | | dsRNA bacteriophage phi6 nucleocapsid | | Descriptor: | Major inner protein P1, Major outer capsid protein, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S.L, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
2VRF
 
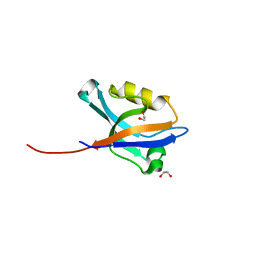 | | CRYSTAL STRUCTURE OF THE HUMAN BETA-2-SYNTROPHIN PDZ DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, BETA-2-SYNTROPHIN | | Authors: | Sun, Z, Roos, A.K, Pike, A.C.W, Pilka, E.S, Cooper, C, Elkins, J.M, Murray, J, Arrowsmith, C.H, Doyle, D, Edwards, A, von Delft, F, Bountra, C, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Beta-2-Syntrophin Pdz Domain
To be Published
|
|
6Q1L
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
1ZVO
 
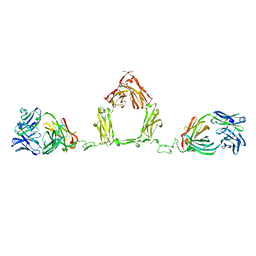 | | Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering | | Descriptor: | Immunoglobulin delta heavy chain, myeloma immunoglobulin D lambda | | Authors: | Sun, Z, Almogren, A, Furtado, P.B, Chowdhury, B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2005-06-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-extended Solution Structure of Human Myeloma Immunoglobulin D Determined by Constrained X-ray Scattering.
J.Mol.Biol., 353, 2005
|
|
1W0S
 
 | |
1W0R
 
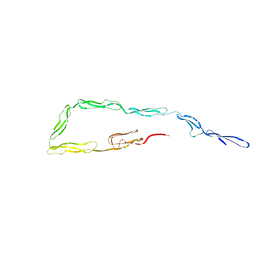 | |
8HR2
 
 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
6Q1B
 
 | |
6PZ0
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and L-Tyrosine | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, TYROSINE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-07-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
8TMR
 
 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMT
 
 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TN0
 
 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
6TYK
 
 | |
7X6W
 
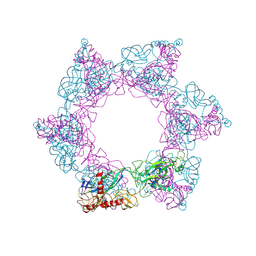 | | SFTSV 2 fold hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (5.18 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X72
 
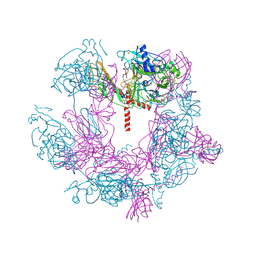 | | SFTSV 5 fold pentamer | | Descriptor: | Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X6U
 
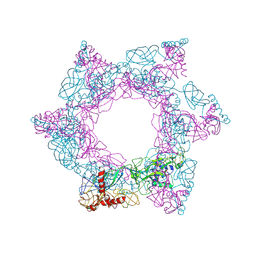 | | SFTSV 3 fold hexmer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
4XBX
 
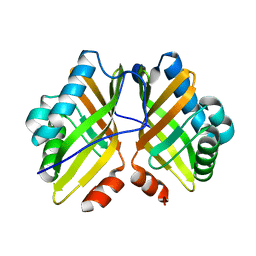 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBT
 
 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH complexed with (S,S)-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CITRATE ANION, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDV
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBY
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDW
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6L62
 
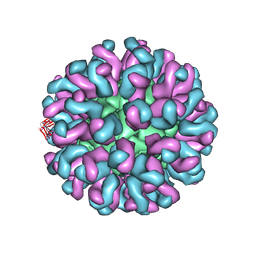 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
