1B0Z
 
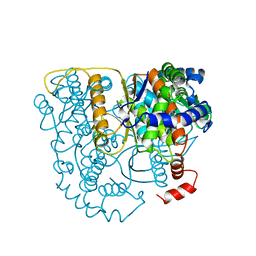 | | The crystal structure of phosphoglucose isomerase-an enzyme with autocrine motility factor activity in tumor cells | | Descriptor: | PROTEIN (PHOSPHOGLUCOSE ISOMERASE) | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-11-15 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition
J.Biol.Chem., 275, 2000
|
|
2BO3
 
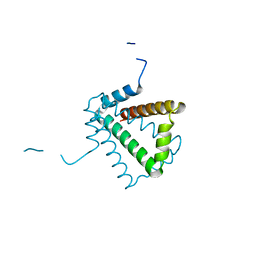 | | Crystal Structure of HP0242, a Hypothetical Protein from Helicobacter pylori | | Descriptor: | HYPOTHETICAL PROTEIN HP0242 | | Authors: | Sun, Y.-J, Tsai, J.-Y, Chen, B.-T. | | Deposit date: | 2005-04-07 | | Release date: | 2006-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Hp0242, a Hypothetical Protein from Helicobacter Pylori with a Novel Fold
Proteins: Struct., Funct., Bioinf., 62, 2006
|
|
2CMH
 
 | |
2CMG
 
 | |
1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
2VW9
 
 | | Single stranded DNA binding protein complex from Helicobacter pylori | | Descriptor: | POLY-DT, SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Chan, K.-W, Wang, C.-H, Lee, Y.-J, Sun, Y.-J. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single-Stranded DNA-Binding Protein Complex from Helicobacter Pylori Suggests an Ssdna-Binding Surface.
J.Mol.Biol., 388, 2009
|
|
2JGQ
 
 | | Kinetics and structural properties of triosephosphate isomerase from Helicobacter pylori | | Descriptor: | 1-[(3-CYCLOHEXYLPROPANOYL)(2-HYDROXYETHYL)AMINO]-1-DEOXY-D-ALLITOL, PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Chu, C.-H, Lai, Y.-J, Sun, Y.-J. | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetics and Structural Properties of Triosephosphate Isomerase from Helicobacter Pylori
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1UVB
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
1UVA
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | MYRISTIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN 1 | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
1UVC
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, STEARIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
6L85
 
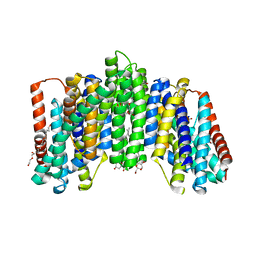 | | The sodium-dependent phosphate transporter | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Sun, Y.-J. | | Deposit date: | 2019-11-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of the sodium-dependent phosphate transporter reveals insights into human solute carrier SLC20.
Sci Adv, 6, 2020
|
|
2WFK
 
 | | Calcium bound LipL32 | | Descriptor: | CALCIUM ION, LIPL32 | | Authors: | Tung, J.-Y, Yang, C.-W, Sun, Y.-J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Binds to Lipl32, a Lipoprotein from Pathogenic Leptospira, and Modulates Fibronectin Binding.
J.Biol.Chem., 285, 2010
|
|
2VQ4
 
 | |
2WIC
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GMPPNP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WIA
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in Apo Form | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WPV
 
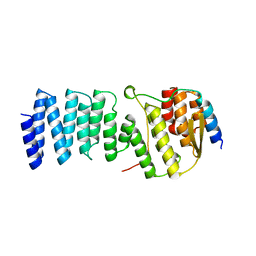 | | Crystal structure of S. cerevisiae Get4-Get5 complex | | Descriptor: | MERCURY (II) ION, UBIQUITIN-LIKE PROTEIN MDY2, UPF0363 PROTEIN YOR164C | | Authors: | Chang, Y.-W, Chuang, Y.-C, Ho, Y.-C, Cheng, M.-Y, Sun, Y.-J, Hsiao, C.-D, Wang, C. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-26 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Get4-Get5 Complex and its Interactions with Sgt2, Get3, and Ydj1.
J.Biol.Chem., 285, 2010
|
|
2WIB
 
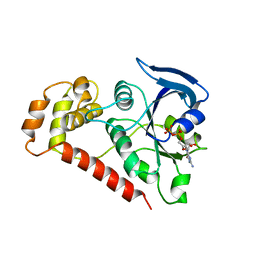 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GDP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2V8L
 
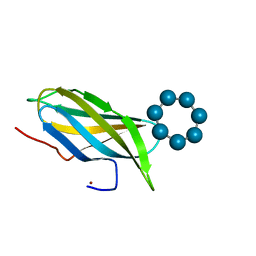 | |
2V8M
 
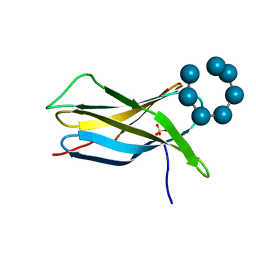 | |
2V7Z
 
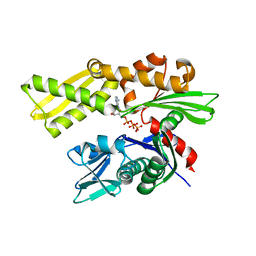 | | Crystal structure of the 70-kDa heat shock cognate protein from Rattus norvegicus in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK COGNATE 71 KDA PROTEIN, PHOSPHATE ION | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
2UZK
 
 | | Crystal structure of the human FOXO3a-DBD bound to DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*AP*G)-3', FORKHEAD BOX PROTEIN O3A | | Authors: | Tsai, K.-L, Sun, Y.-J, Huang, C.-Y, Yang, J.-Y, Hung, M.-C, Hsiao, C.-D. | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Human Foxo3A-Dbd/DNA Complex Suggests the Effects of Post-Translational Modification.
Nucleic Acids Res., 35, 2007
|
|
2V7Y
 
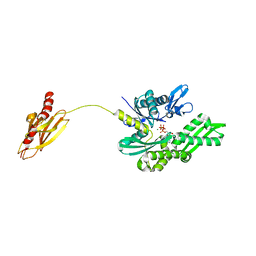 | | Crystal structure of the molecular chaperone DnaK from Geobacillus kaustophilus HTA426 in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN DNAK, MAGNESIUM ION, ... | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
1OJ1
 
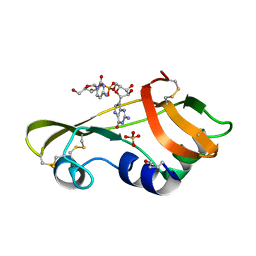 | | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with (2,5 CpG) and d(ApCpGpA) | | Descriptor: | CYTIDYL-2'-5'-PHOSPHO-GUANOSINE, RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-06-28 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with C(2,5 Cpg) and D(Apcpgpa)
To be Published
|
|
1OJ8
 
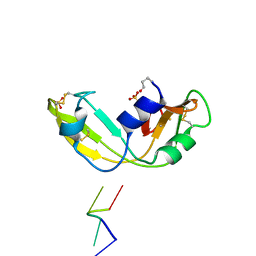 | | Novel and retro Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with d(ApCpGpA) and (2',5'CpG) | | Descriptor: | 5'-D(*AP*CP*GP*AP)-3', RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-Y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-07-07 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retro and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with (2',5'Cpg) and D(Apcpgpa)
To be Published
|
|
