3GGO
 
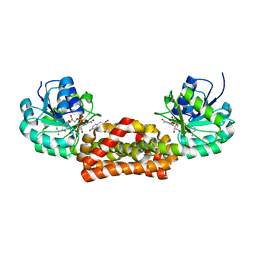 | | Crystal structure of prephenate dehydrogenase from A. aeolicus with HPP and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Prephenate dehydrogenase | | Authors: | Sun, W, Shahinas, D, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3GGP
 
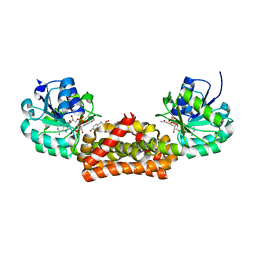 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3GGG
 
 | |
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | Descriptor: | hypothetical protein PH1915 | | Authors: | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
2G5C
 
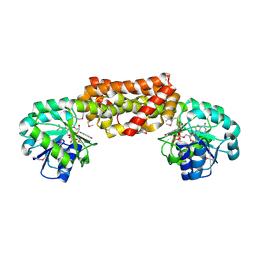 | | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, prephenate dehydrogenase | | Authors: | Sun, W, Singh, S, Zhang, R, Turnbull, J.L, Christendat, D. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus: Insights into the Catalytic Mechanism
J.Biol.Chem., 281, 2006
|
|
4KK1
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
4KK0
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
5YDE
 
 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.023 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
5YDF
 
 | | Crystal structure of a disease-related gene, hCDC73(1-100) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
8HNT
 
 | |
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNW
 
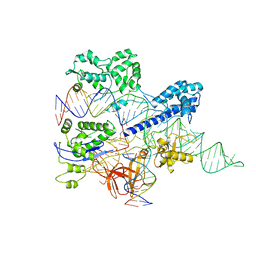 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNV
 
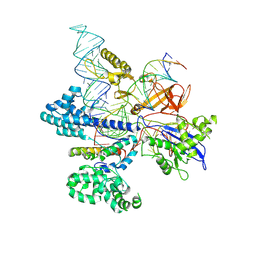 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | Descriptor: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
7D68
 
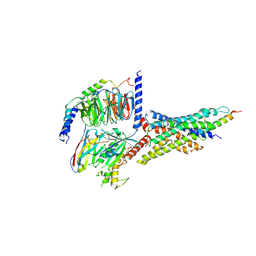 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
7YBF
 
 | |
4IC6
 
 | | Crystal structure of Deg8 | | Descriptor: | Protease Do-like 8, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IC5
 
 | | Crystal structure of Deg5 | | Descriptor: | CALCIUM ION, Protease Do-like 5, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
