1AA6
 
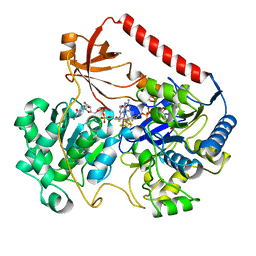 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1FDO
 
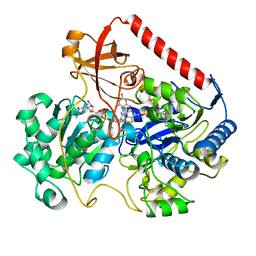 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1FDI
 
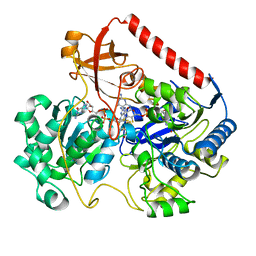 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI COMPLEXED WITH THE INHIBITOR NITRITE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
2DLI
 
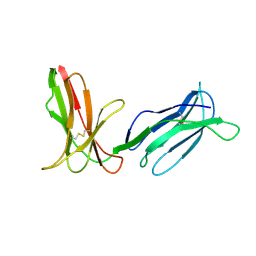 | | KILLER IMMUNOGLOBULIN RECEPTOR 2DL2,TRIGONAL FORM | | Descriptor: | PROTEIN (MHC CLASS I NK CELL RECEPTOR PRECURSOR) | | Authors: | Sun, P.D, Snyder, G.A. | | Deposit date: | 1999-03-08 | | Release date: | 2000-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6ULR
 
 | |
6ULI
 
 | |
6ULK
 
 | |
6ULN
 
 | |
6UON
 
 | |
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.485 Å) | | Cite: | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1EFX
 
 | | STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA-CW3 (HEAVY CHAIN), NATURAL KILLER CELL RECEPTOR KIR2DL2, ... | | Authors: | Boyington, J.C, Motyka, S.A, Schuck, P, Brooks, A.G, Sun, P.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand.
Nature, 405, 2000
|
|
1B6E
 
 | | HUMAN CD94 | | Descriptor: | CD94 | | Authors: | Boyington, J.C, Riaz, A.N, Patamawenu, A, Coligan, J.E, Brooks, A.G, Sun, P.D. | | Deposit date: | 1999-01-14 | | Release date: | 1999-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors.
Immunity, 10, 1999
|
|
2ZG2
 
 | |
2ZG1
 
 | |
2ZG3
 
 | |
1XPH
 
 | |
2AU1
 
 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | Descriptor: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-26 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2AVW
 
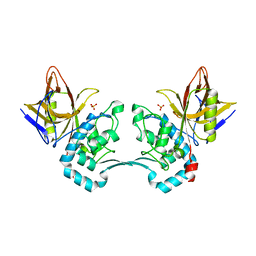 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
1FNL
 
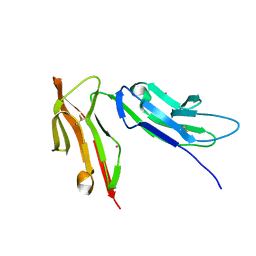 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF A HUMAN FCGRIII | | Descriptor: | LOW AFFINITY IMMUNOGLOBULIN GAMMA FC REGION RECEPTOR III-B, MERCURY (II) ION | | Authors: | Zhang, Y, Boesen, C.C, Radaev, S, Brooks, A.G, Fridman, W.H, Sautes-Fridman, C, Sun, P.D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the extracellular domain of a human Fc gamma RIII.
Immunity, 13, 2000
|
|
8DIR
 
 | |
8DJ7
 
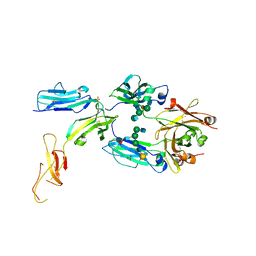 | | The complex structure between human IgG1 Fc and its high affinity receptor FcgRI H174R variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin gamma Fc receptor I, Ig gamma-1 Fc chain, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fc gamma RI FG-loop functions as a pH sensitive switch for IgG binding and release.
Front Immunol, 14, 2023
|
|
8DIN
 
 | |
1P6F
 
 | | Structure of the human natural cytotoxicity receptor NKp46 | | Descriptor: | natural cytotoxicity triggering receptor 1 | | Authors: | Foster, C.E, Colonna, M, Sun, P.D. | | Deposit date: | 2003-04-29 | | Release date: | 2003-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors.
J.Biol.Chem., 278, 2003
|
|
1Q8M
 
 | | Crystal structure of the human myeloid cell activating receptor TREM-1 | | Descriptor: | GLUTATHIONE, SULFATE ION, triggering receptor expressed on myeloid cells 1 | | Authors: | Radaev, S, Kattah, M, Rostro, B, Colonna, M, Sun, P.D. | | Deposit date: | 2003-08-21 | | Release date: | 2003-12-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human myeloid cell activating receptor TREM-1
Structure, 11, 2003
|
|
1M9Z
 
 | | CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN | | Descriptor: | GLYCEROL, TGF-BETA RECEPTOR TYPE II | | Authors: | Boesen, C.C, Radaev, S, Motyka, S.A, Patamawenu, A, Sun, P.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | THE 1.1A CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN
Structure, 10, 2002
|
|
