5HLJ
 
 | | Crystal Structure of Major Envelope Protein VP24 from White Spot Syndrome Virus | | Descriptor: | VP24 | | Authors: | Sun, L.F, Su, Y.T, Zhao, Y.H, Fu, Z.Q, Wu, Y.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Crystal Structure of Major Envelope Protein VP24 from White Spot Syndrome Virus
Sci Rep, 6, 2016
|
|
5ZKT
 
 | |
7VIV
 
 | |
3O7Q
 
 | | Crystal structure of a Major Facilitator Superfamily (MFS) transporter, FucP, in the outward conformation | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Dang, S.Y, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.143 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
4NUR
 
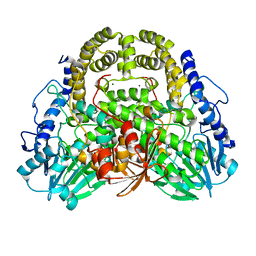 | | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9 | | Descriptor: | MAGNESIUM ION, PSdsA, ZINC ION | | Authors: | Sun, L.F, Su, Y.T, Zhao, Y.H, Cai, Z.X, Wu, Y. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Crystal structure of thermostable alkylsulfatase SdsAP from Pseudomonas sp. S9
To be Published
|
|
3O7P
 
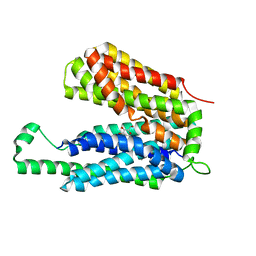 | | Crystal structure of the E.coli Fucose:proton symporter, FucP (N162A) | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Dang, S.Y, Sun, L.F, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
6KSW
 
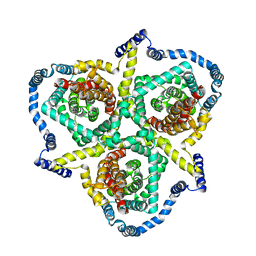 | | Cryo-EM structure of the human concentrative nucleoside transporter CNT3 | | Descriptor: | Solute carrier family 28 member 3 | | Authors: | Zhou, Y.X, Liao, L.H, Li, J.L, Xiao, Q.J, Sun, L.F, Deng, D. | | Deposit date: | 2019-08-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the human concentrative nucleoside transporter CNT3.
Plos Biol., 18, 2020
|
|
6KXT
 
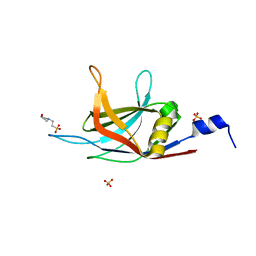 | | BON1-C2B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXU
 
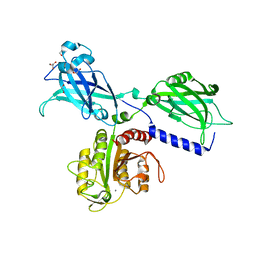 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXK
 
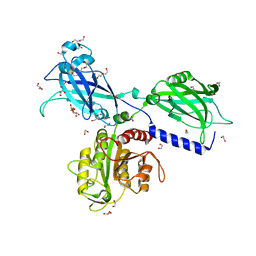 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
4XH3
 
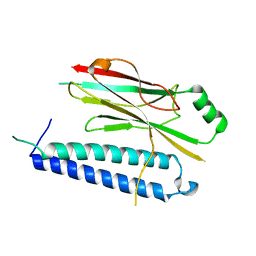 | |
4GBY
 
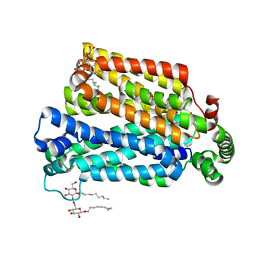 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
7XHN
 
 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
5GY3
 
 | |
4GC0
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4O8S
 
 | |
7XHO
 
 | | Structure of human inner kinetochore CCAN complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
6J4R
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J4K
 
 | |
6J5B
 
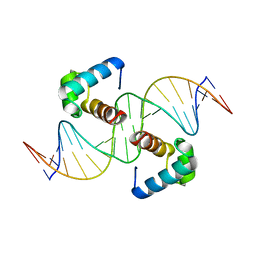 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6JBH
 
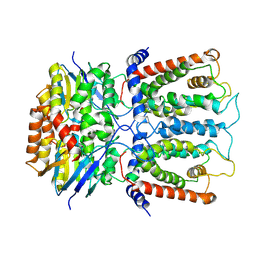 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
6JBJ
 
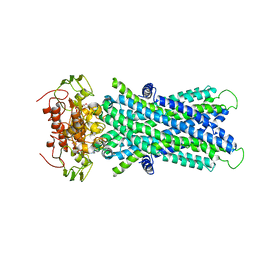 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
6L30
 
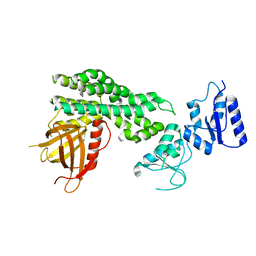 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LR0
 
 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
