3OPY
 
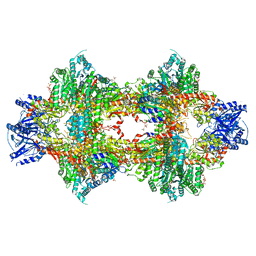 | | Crystal structure of Pichia pastoris phosphofructokinase in the T-state | | Descriptor: | 6-phosphofructo-1-kinase alpha-subunit, 6-phosphofructo-1-kinase beta-subunit, 6-phosphofructo-1-kinase gamma-subunit, ... | | Authors: | Strater, N, Marek, S, Kuettner, E.B, Kloos, M, Keim, A, Bruser, A, Kirchberger, J, Schoneberg, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Faseb J., 25, 2011
|
|
8Q71
 
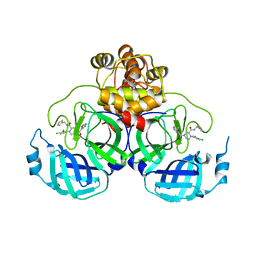 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
6Z9B
 
 | |
6Z9D
 
 | |
8C9W
 
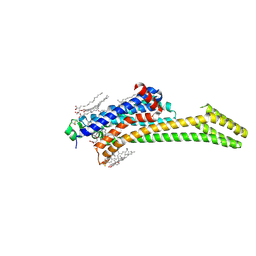 | | Crystal structure of the adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with Etrumadenant at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, ... | | Authors: | Strater, N, Claff, T, Schlegel, J.G, Voss, J.H, Vaassen, V, Muller, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Crystal structure of adenosine A 2A receptor in complex with clinical candidate Etrumadenant reveals unprecedented antagonist interaction.
Commun Chem, 6, 2023
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
7QGA
 
 | |
7QGM
 
 | |
7QGO
 
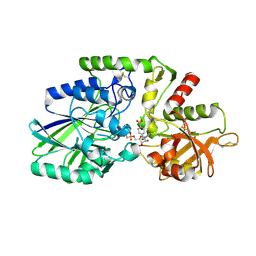 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QGL
 
 | |
6XUE
 
 | |
6XUG
 
 | |
6XUQ
 
 | |
5EI0
 
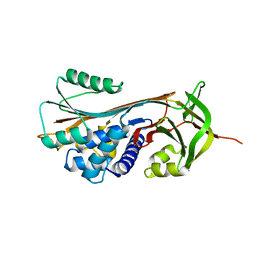 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
4WL0
 
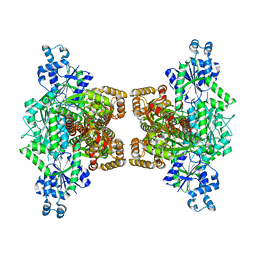 | |
2VOA
 
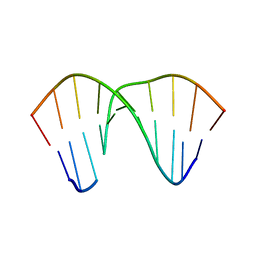 | | Structure of an AP Endonuclease from Archaeoglobus fulgidus | | Descriptor: | 5'-D(*CP*GP*GP*CP*TP*AP*CP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*GP*TP*AP*GP*CP*CP*GP)-3', EXODEOXYRIBONUCLEASE III | | Authors: | Kuettner, E.B, Schmiedel, R, Greiner-Stoffele, T, Strater, N. | | Deposit date: | 2008-02-11 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Abasic Site Specificity Pocket of an Ap Endonuclease from Archaeoglobus Fulgidus.
DNA Repair, 8, 2009
|
|
8OEK
 
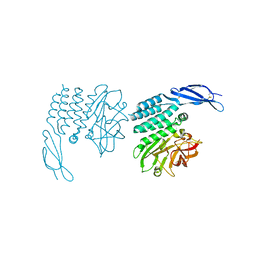 | |
7NEI
 
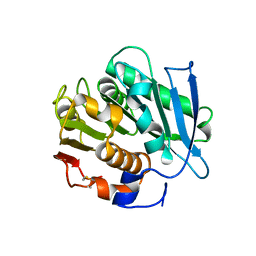 | |
4WWL
 
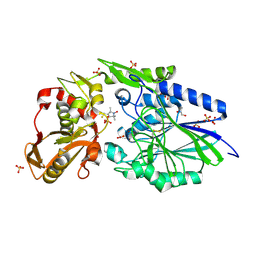 | | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form) | | Descriptor: | CARBONATE ION, GLYCEROL, Protein UshA, ... | | Authors: | Krug, U, Paithankar, K.S, Schultz-Heienbrok, R, Strater, N. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form)
To Be Published
|
|
3KBP
 
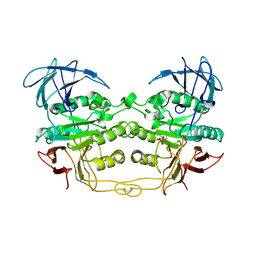 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-10-02 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
1O7A
 
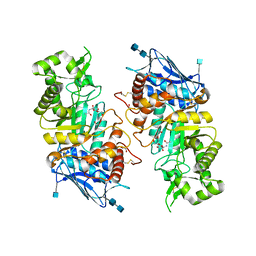 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
1KBP
 
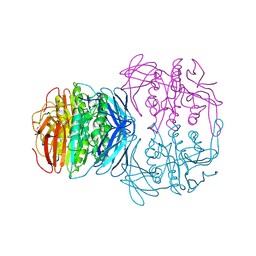 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
6HDX
 
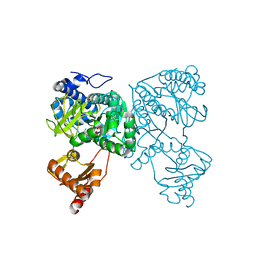 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE2
 
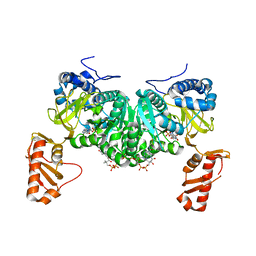 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
