2WJ3
 
 | |
2WJ6
 
 | |
2WJ4
 
 | |
2WM2
 
 | |
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GQH
 
 | | Quercetin 2,3-dioxygenase in complex with the inhibitor kojic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
1H1M
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE KAEMPFEROL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GQG
 
 | | Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
1H1I
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE QUERCETN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-15 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2XG7
 
 | | Crystal Structure of BST2-Tetherin Ectodomain expressed in HEK293T cells | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BONE MARROW STROMAL ANTIGEN 2 | | Authors: | Steiner, R.A. | | Deposit date: | 2010-05-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural and Functional Studies on the Extracellular Domain of Bst2/Tetherin in Reduced and Oxidized Conformations.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8OIH
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under oxidising conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, BROMIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
8OFK
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
4UT2
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with ascorbate | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Kopec, J, Zeh Silva, M, Fotinou, C, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
7Q2T
 
 | |
7QAR
 
 | | Serial crystallography structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-methyl uric acid at room temperature | | Descriptor: | (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, Uricase | | Authors: | Bui, S, Catapano, L, Zielinski, K, Yefanov, O, Murshudov, G.N, Oberthuer, D, Steiner, R.A. | | Deposit date: | 2021-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
6FUZ
 
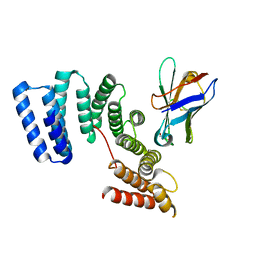 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of JIP1 | | Descriptor: | GLYCEROL, Kinesin light chain 1,Kinesin light chain 1,C-Jun-amino-terminal kinase-interacting protein 1, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
6FV0
 
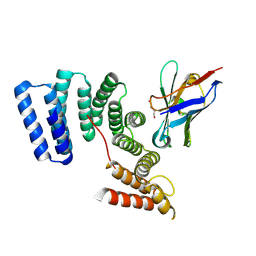 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of torsinA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Kinesin light chain 1,Torsin-1A, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
8OXN
 
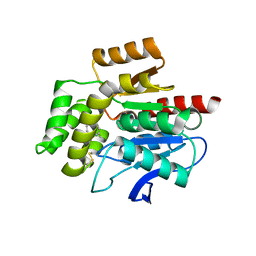 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8ORO
 
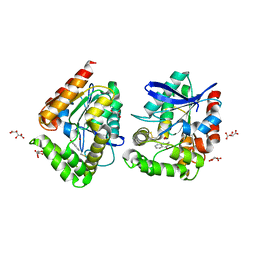 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER HYPEROXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-04-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8OH8
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group P 21 21 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
8OXT
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) H251A VARIANT COMPLEXED WITH N-ACETYLANTHRANILATE AS RESULT OF IN CRYSTALLO TURNOVER OF ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-(ACETYLAMINO)BENZOIC ACID, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8OIW
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under oxidising conditions (space group P 21 21 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
4UOW
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
4UT3
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with hydrogen peroxide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Zeh Silva, M, Kopec, J, Fotinou, D, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
7OKZ
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL- QUINOLIN-4(1H)-ONE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
