443D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'/ BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
1X8B
 
 | | Structure of human Wee1A kinase: kinase domain complexed with inhibitor PD0407824 | | Descriptor: | 9-HYDROXY-4-PHENYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, MAGNESIUM ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2004-08-17 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and inhibition of the human cell cycle checkpoint kinase, Wee1A kinase: an atypical tyrosine kinase with a key role in CDK1 regulation
Structure, 13, 2005
|
|
4DZ5
 
 | |
4WRG
 
 | |
447D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-18 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
449D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
445D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', Benzimidazole derivative complex | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
3CQE
 
 | | Wee1 kinase complex with inhibitor PD074291 | | Descriptor: | 8-bromo-4-(2-chlorophenyl)-N-(2-hydroxyethyl)-6-methyl-1,3-dioxo-1,2,3,6-tetrahydropyrrolo[3,4-e]indole-7-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinants of Wee1 Inhibitor Selectivity
To be Published
|
|
3CR0
 
 | | Wee1 kinase complex with inhibitor PD259_809 | | Descriptor: | 4-(2-chlorophenyl)-8-(2-hydroxyethyl)-6-methylpyrrolo[3,4-e]indole-1,3(2H,6H)-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2008-04-03 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of Wee1 inhibitor selectivity
To be Published
|
|
2Z2W
 
 | | Human Wee1 kinase complexed with inhibitor PF0335770 | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-(2-CHLOROPHENYL)-1,3-DIOXO-1,2,3,6-TETRAHYDROPYRROLO[3,4-C]CARBAZOL-9-YL]FORMAMIDE, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2007-05-29 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 9-Amino-4-(2-chlorophenyl)pyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones and Related Formamides as Inhibitors of the Wee1 and Chk1 Checkpoint Kinases
To be Published
|
|
328D
 
 | | STRUCTURE OF A D(CGCGAATTCGCG)2-SN7167 COMPLEX | | Descriptor: | 4-[4-[2-AMINO-4-[4,6-(N-METHYLQUINOLINIUM)AMINO]BENZAMIDO]ANILINO]-N-METHYLPYRIDINIUM MESYLATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Clark, G.R, Denny, W.A. | | Deposit date: | 1997-04-15 | | Release date: | 1997-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Minor groove binding of a bis-quaternary ammonium compound: the crystal structure of SN 7167 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 25, 1997
|
|
4WUN
 
 | | Structure of FGFR1 in complex with AZD4547 (N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE) at 1.65 angstrom | | Descriptor: | Fibroblast growth factor receptor 1, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Squire, C.J, Yosaatmadja, C.J. | | Deposit date: | 2014-11-02 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The 1.65 angstrom resolution structure of the complex of AZD4547 with the kinase domain of FGFR1 displays exquisite molecular recognition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2IO6
 
 | | Wee1 kinase complexed with inhibitor PD330961 | | Descriptor: | 9-HYDROXY-6-(3-HYDROXYPROPYL)-4-(2-METHOXYPHENYL)PYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
2IN6
 
 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4WRH
 
 | | AKR1C3 complexed with breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(aminomethyl)-2-chloro-6-methoxyphenoxy]-N-tert-butylacetamide, 5-methyl-4H-1,2,4-triazole-3-thiol, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide trapped in active site of AKR1C3
To Be Published
|
|
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
1QV4
 
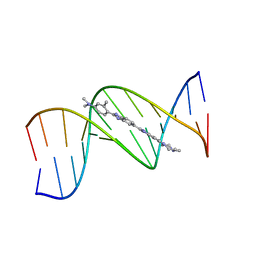 | |
1QV8
 
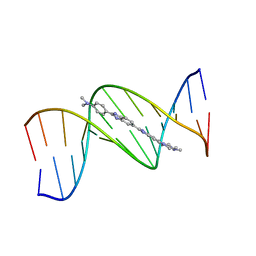 | | B-DNA Dodecamer d(CGCGAATTCGCG)2 complexed with proamine | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Squire, C.J, Clark, G.R. | | Deposit date: | 2003-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In vitro studies with methylproamine: a potent new radioprotector.
Cancer Res., 64, 2004
|
|
4ZAU
 
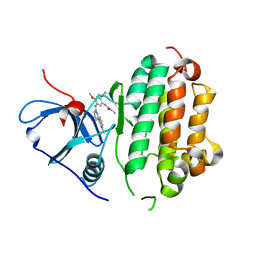 | | AZD9291 complex with wild type EGFR | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Squire, C.J, Yosaatmadja, Y, Flanagan, J.U, McKeage, M. | | Deposit date: | 2015-04-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed.
J.Struct.Biol., 192, 2015
|
|
444D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
442D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
448D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
3BI6
 
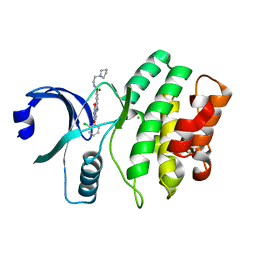 | | Wee1 kinase complex with inhibitor PD352396 | | Descriptor: | 4-(2-chlorophenyl)-9-hydroxy-6-methyl-1,3-dioxo-N-(2-pyrrolidin-1-ylethyl)pyrrolo[3,4-g]carbazole-8-carboxamide, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BIZ
 
 | | Wee1 kinase complex with inhibitor PD331618 | | Descriptor: | 4-(2-chlorophenyl)-8-[3-(dimethylamino)propoxy]-9-hydroxy-6-methylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-dione, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-12-02 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
