5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
7ZJX
 
 | | Rabbit 80S ribosome programmed with SECIS and SBP2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | Authors: | Hilal, T, Simonovic, M, Spahn, C.M.T. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
7ZJW
 
 | | Rabbit 80S ribosome as it decodes the Sec-UGA codon | | Descriptor: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | Authors: | Hilal, T, Simonovic, M, Spahn, C.M.T. | | Deposit date: | 2022-04-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
3DEG
 
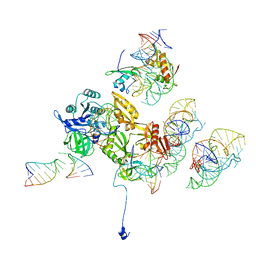 | | Complex of elongating Escherichia coli 70S ribosome and EF4(LepA)-GMPPNP | | Descriptor: | 30S RNA helix 14, 30S RNA helix 8, 30S ribosomal protein S12, ... | | Authors: | Connell, S.R, Topf, M, Qin, Y, Wilson, D.N, Mielke, T, Fucini, P, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation
Nat.Struct.Mol.Biol., 15, 2008
|
|
8B2L
 
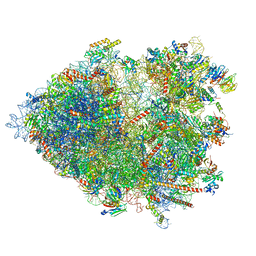 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
2IWH
 
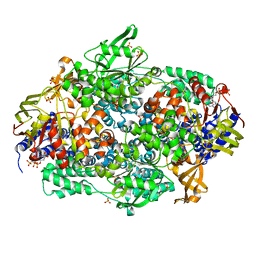 | | Structure of yeast Elongation Factor 3 in complex with ADPNP | | Descriptor: | ELONGATION FACTOR 3A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IX8
 
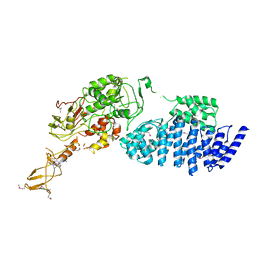 | | MODEL FOR EEF3 BOUND TO AN 80S RIBOSOME | | Descriptor: | ELONGATION FACTOR 3A | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-10 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IW3
 
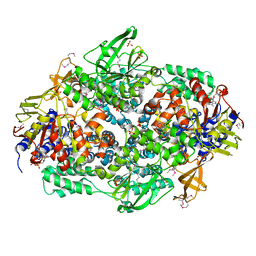 | | Elongation Factor 3 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELONGATION FACTOR 3A, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IX3
 
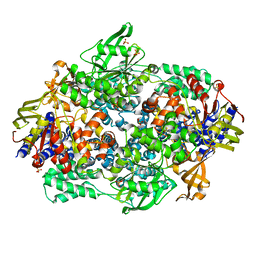 | | Structure of yeast Elongation Factor 3 | | Descriptor: | ELONGATION FACTOR 3, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
4V5M
 
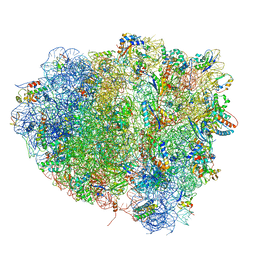 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4V5N
 
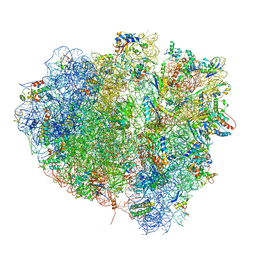 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
7BL5
 
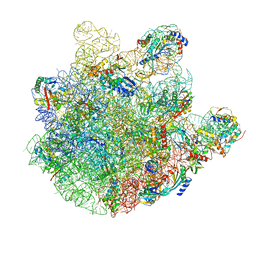 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL2
 
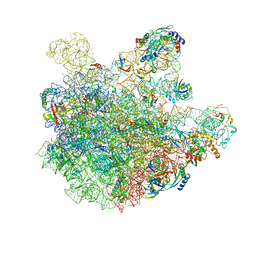 | | pre-50S-ObgE particle state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL4
 
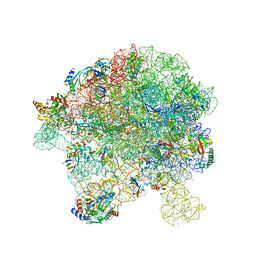 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL6
 
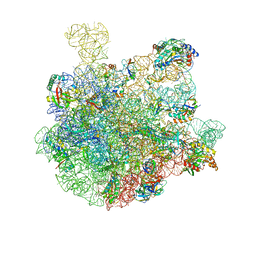 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL3
 
 | | pre-50S-ObgE particle state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UJC
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6YGC
 
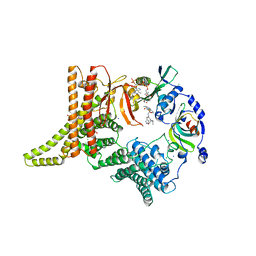 | | Crystal structure of the NatC complex bound to Arl3 peptide and CoA | | Descriptor: | ADP-ribosylation factor-like protein 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YGB
 
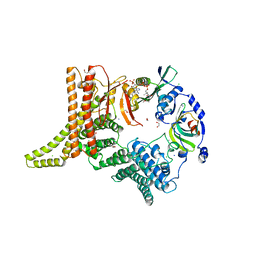 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YGA
 
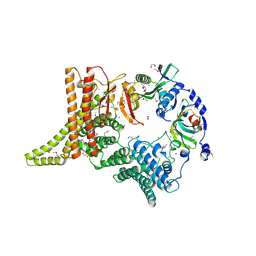 | | Crystal structure of the apo NatC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YGD
 
 | | Crystal structure of the NatC complex bound to Gag peptide and CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
4D67
 
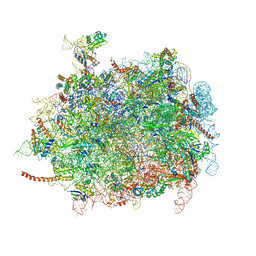 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 28S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-04 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em Structures of Ribosomal 80S Complexes with Termination Factors and Cricket Paralysis Virus Ires Reveal the Ires in the Translocated State
Mol.Cell, 57, 2015
|
|
4D61
 
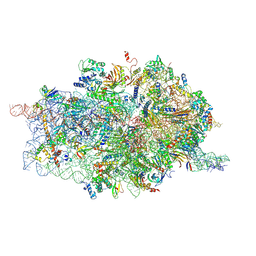 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5N
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | CRICKET PARALYSIS VIRUS IRES RNA, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1 | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
