6F3K
 
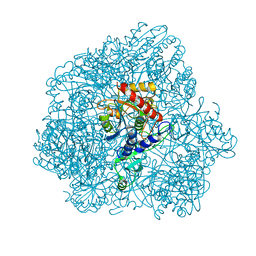 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6G7O
 
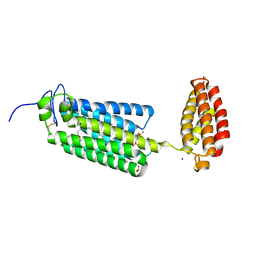 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
7R0J
 
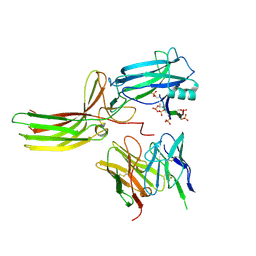 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
7R0C
 
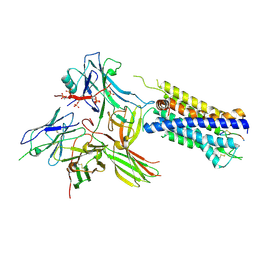 | | Structure of the AVP-V2R-arrestin2-ScFv30 complex | | Descriptor: | AVP, Arrestin2, ScFv30, ... | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
7BB7
 
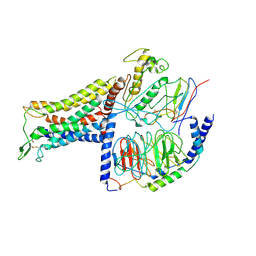 | | AVP-V2R-Galphas-beta1-gamma2-Nb35(T state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Bous, J, Mouillac, B, Bron, P, Granier, S, Floquet, N, Leyrat, C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-electron microscopy structure of the antidiuretic hormone arginine-vasopressin V2 receptor signaling complex.
Sci Adv, 7, 2021
|
|
7BB6
 
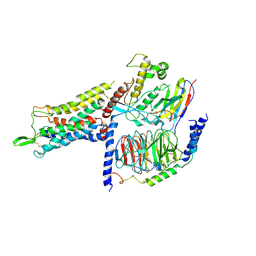 | | AVP-V2R-Galphas-beta1-gamma2-Nb35 (L state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Bous, J, Mouillac, B, Bron, P, Granier, S, Floquet, N, Leyrat, C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structure of the antidiuretic hormone arginine-vasopressin V2 receptor signaling complex.
Sci Adv, 7, 2021
|
|
5LX9
 
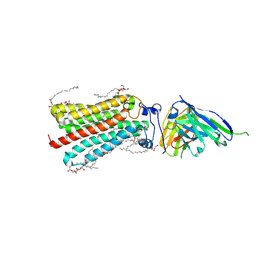 | | CRYSTAL STRUCTURE OF HUMAN ADIPONECTIN RECEPTOR 2 IN COMPLEX WITH A C18 FREE FATTY ACID AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, HUMAN ADIPONECTIN RECEPTOR 2, OLEIC ACID, ... | | Authors: | Vasiliauskaite-Brooks, I, Leyrat, C, Hoh, F, Granier, S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-22 | | Last modified: | 2017-04-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
5LXG
 
 | | Revised crystal structure of the human adiponectin receptor 1 in an open conformation | | Descriptor: | Adiponectin receptor protein 1, SULFATE ION, V REGION HEAVY CHAIN, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Granier, S. | | Deposit date: | 2016-09-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
5LXA
 
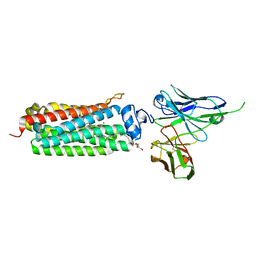 | | CRYSTAL STRUCTURE OF HUMAN ADIPONECTIN RECEPTOR 2 IN COMPLEX WITH A C18 FREE FATTY ACID AT 3.0 ANGSTROM RESOLUTION | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, Anti-CD30 moab Ki-4 scFv, ... | | Authors: | Vasiliauskaite-Brooks, I, Leyrat, C, Hoh, F, Granier, S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
5LWY
 
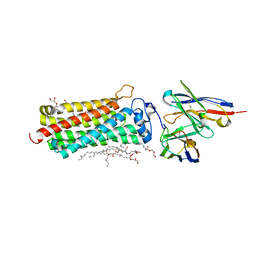 | | Revised crystal structure of the human adiponectin receptor 2 in complex with a C18 free fatty acid | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Granier, S. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
