1KDE
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, 22 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1KDF
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
2AFP
 
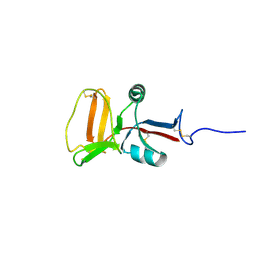 | | THE SOLUTION STRUCTURE OF TYPE II ANTIFREEZE PROTEIN REVEALS A NEW MEMBER OF THE LECTIN FAMILY | | Descriptor: | PROTEIN (SEA RAVEN TYPE II ANTIFREEZE PROTEIN) | | Authors: | Gronwald, W, Loewen, M.C, Lix, B, Daugulis, A.J, Sonnichsen, F.D, Davies, P.L, Sykes, B.D. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of type II antifreeze protein reveals a new member of the lectin family.
Biochemistry, 37, 1998
|
|
2K21
 
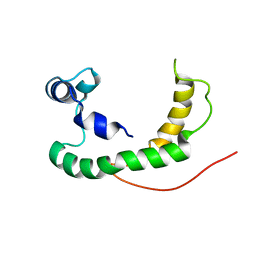 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2JS2
 
 | |
2JS0
 
 | |
2K1D
 
 | |
1DD2
 
 | |
1DCZ
 
 | |
1PAK
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
1TRF
 
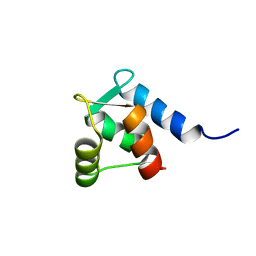 | |
3AME
 
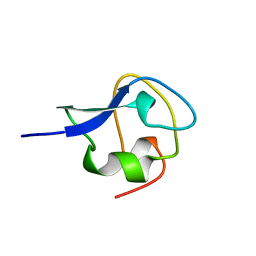 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 Q9TQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1C3Y
 
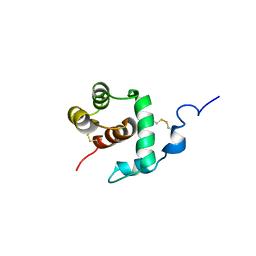 | | THP12-CARRIER PROTEIN FROM YELLOW MEAL WORM | | Descriptor: | THP12 CARRIER PROTEIN | | Authors: | Soennichsen, F.D. | | Deposit date: | 1999-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands.
Structure Fold.Des., 7, 1999
|
|
4AME
 
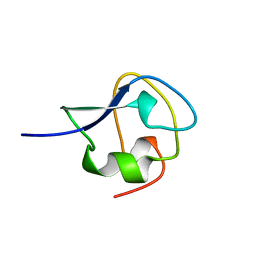 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3NLA
 
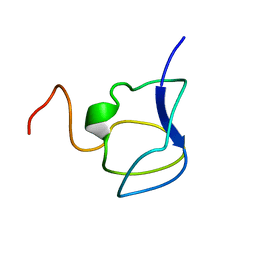 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
7AME
 
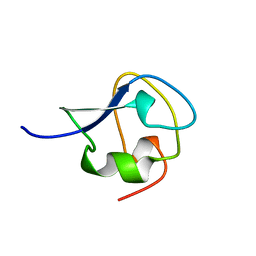 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
2LN8
 
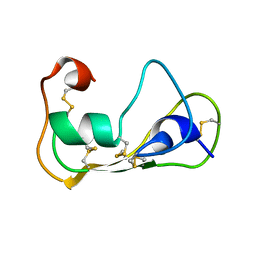 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
2LRD
 
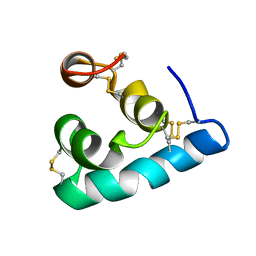 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LXO
 
 | | Identification of the Structural Traits Mediating the Antimicrobial Activity of a Chimeric Peptide of HBD2 and HBD3 | | Descriptor: | Chimeric Peptide | | Authors: | Spudy, B, Soennichsen, F.D, Waetzig, G.H, Grotzinger, J, Jung, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-13 | | Method: | SOLUTION NMR | | Cite: | Identification of structural traits that increase the antimicrobial activity of a chimeric peptide of human beta-defensins 2 and 3.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
2M2F
 
 | | The membran-proximal domain of ADAM17 | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17 | | Authors: | Duesterhoeft, S, Jung, S, Hung, C, Tholey, A, Soennichsen, F.D, Groetzinger, J, Lorenzen, I. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase.
J.Am.Chem.Soc., 135, 2013
|
|
2M8T
 
 | |
2MXQ
 
 | | The solution structure of DEFA1, a highly potent antimicrobial peptide from the horse | | Descriptor: | Paneth cell-specific alpha-defensin 1 | | Authors: | Jung, S, Michalek, M, Shomali, M, Soennichsen, F.D. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional studies of the highly potent equine antimicrobial peptide DEFA1.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
