1PIC
 
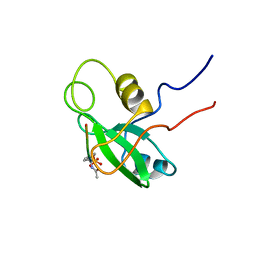 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Breeze, A.L, Kara, B.V, Barratt, D.G, Anderson, M, Smith, J.C, Luke, R.W, Best, J.R, Cartlidge, S.A. | | Deposit date: | 1997-06-23 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a specific peptide complex of the carboxy-terminal SH2 domain from the p85 alpha subunit of phosphatidylinositol 3-kinase.
EMBO J., 15, 1996
|
|
7S3K
 
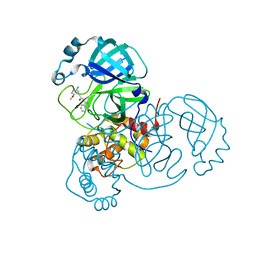 | |
6ZGC
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | Descriptor: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | Authors: | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
7S4B
 
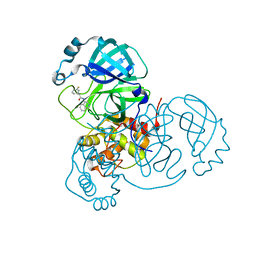 | |
7S3S
 
 | |
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFI
 
 | | SK678 binding region (Siglec + Unique) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFB
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, GLYCEROL, SK150 siglec + Unique, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EF7
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFA
 
 | | GspB Siglec + Unique domains | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EF9
 
 | | GspB Siglec domain | | Descriptor: | FORMIC ACID, Platelet binding protein GspB - Siglec domain, SODIUM ION | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFD
 
 | | Hsa Siglec and Unique domains in complex with the sialyl T antigen trisaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFF
 
 | | NCTC10712 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5K
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6BBS
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, Aggarwal, M, McKenna, R. | | Deposit date: | 2017-10-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BCC
 
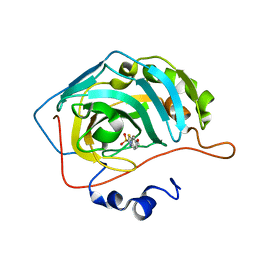 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with ethoxzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BC9
 
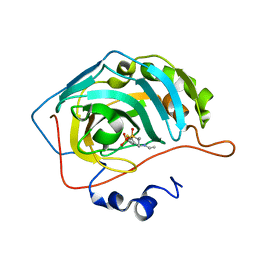 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6X3K
 
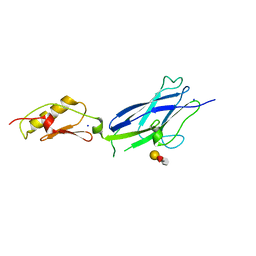 | |
6X3Q
 
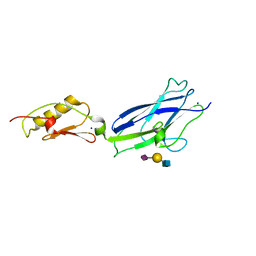 | |
5C8I
 
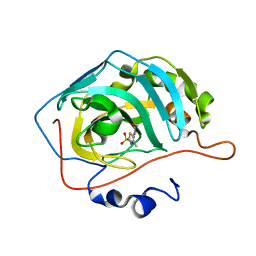 | | Joint X-ray/neutron structure of Human Carbonic Anhydrase II in complex with Methazolamide | | Descriptor: | Carbonic anhydrase 2, N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2(3H)-ylidene)acetamide, ZINC ION | | Authors: | Aggarwal, M, Kovalevsky, A.Y, Fisher, S.Z, McKenna, R. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.56 Å), X-RAY DIFFRACTION | | Cite: | Neutron structure of human carbonic anhydrase II in complex with methazolamide: mapping the solvent and hydrogen-bonding patterns of an effective clinical drug.
IUCrJ, 3, 2016
|
|
4QEE
 
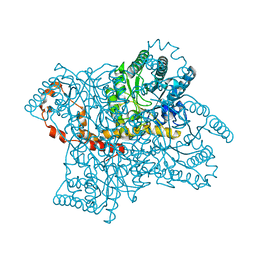 | |
4QDW
 
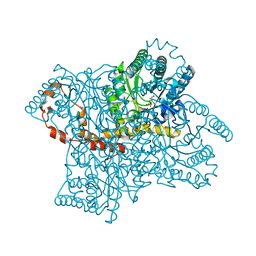 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE5
 
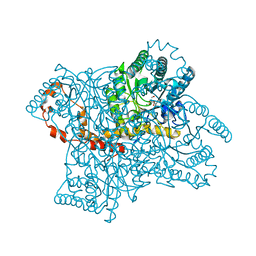 | |
4QE4
 
 | |
