8FAJ
 
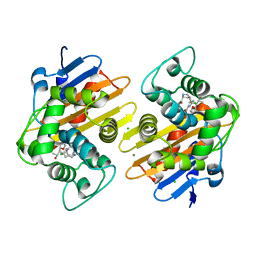 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
4QC6
 
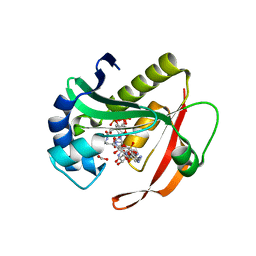 | | Crystal structure of aminoglycoside 6'-acetyltransferase-Ie | | Descriptor: | (3R,5S,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3,5-diphosphaheptadecane-17-sulfinic acid 3,5-dioxide (non-preferred name), Bifunctional AAC/APH, FORMIC ACID, ... | | Authors: | Smith, C.A, Toth, M, Weiss, T.M, Frase, H, Vakulenko, S.B. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the bifunctional aminoglycoside-resistance enzyme AAC(6')-Ie-APH(2'')-Ia revealed by crystallographic and small-angle X-ray scattering analysis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LCF
 
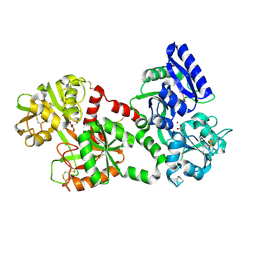 | | CRYSTAL STRUCTURE OF COPPER-AND OXALATE-SUBSTITUTED HUMAN LACTOFERRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1994-01-11 | | Release date: | 1994-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of copper- and oxalate-substituted human lactoferrin at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1MNE
 
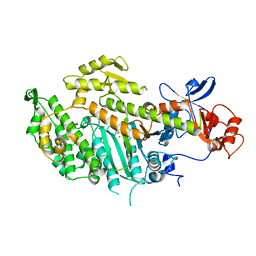 | |
7USZ
 
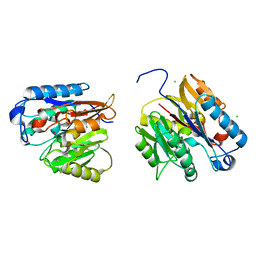 | | Human DDAH-1, holo (Zn-bound) form | | Descriptor: | CHLORIDE ION, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, ZINC ION | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
7UT0
 
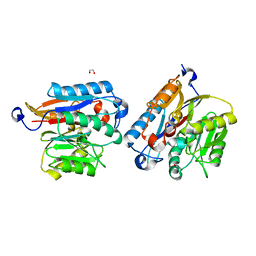 | | Human DDAH-1, apo form | | Descriptor: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
6BFF
 
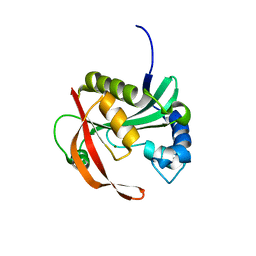 | |
6BFH
 
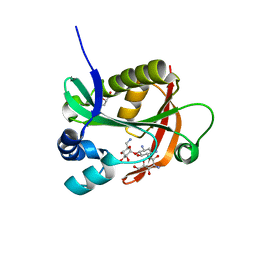 | |
4ZDX
 
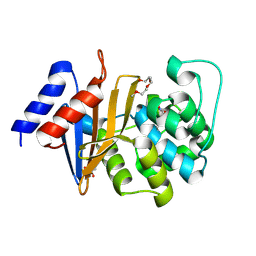 | | Structure of OXA-51 beta-lactamase | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Beta-lactamase, GLYCEROL | | Authors: | Smith, C.A, Antunes, N.T, Stewart, N.K, Frase, H, Toth, M, Kantardjieff, K.A, Vakulenko, S.B. | | Deposit date: | 2015-04-20 | | Release date: | 2015-06-17 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D beta-Lactamases.
Acs Chem.Biol., 10, 2015
|
|
3TDV
 
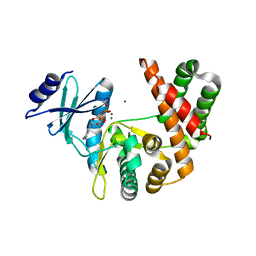 | |
3TDW
 
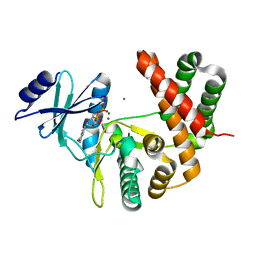 | |
1LFI
 
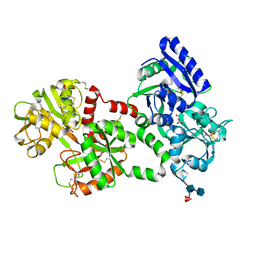 | | METAL SUBSTITUTION IN TRANSFERRINS: THE CRYSTAL STRUCTURE OF HUMAN COPPER-LACTOFERRIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1992-02-10 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metal substitution in transferrins: the crystal structure of human copper-lactoferrin at 2.1-A resolution.
Biochemistry, 31, 1992
|
|
6CTZ
 
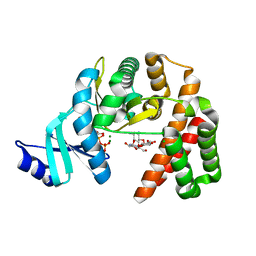 | | Structure of the GDP and kanamycin complex of APH(2")-IIia | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Gentamicin resistance protein, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the diversity of the mechanism of nucleotide hydrolysis by the aminoglycoside-2''-phosphotransferases
Acta Crystallogr.,Sect.D, 75, 2019
|
|
5F83
 
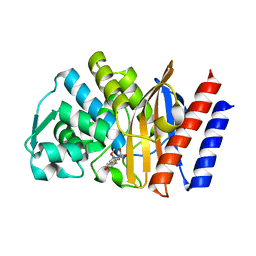 | | Imipenem complex of the GES-5 C69G mutant | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5F82
 
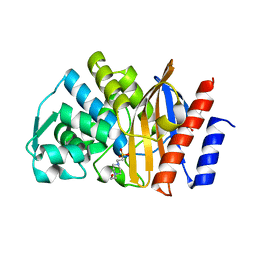 | | Apo GES-5 C69G mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-08 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
6P96
 
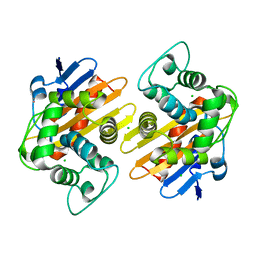 | | OXA-48 carbapanemase, apo form | | Descriptor: | Beta-lactamase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P9C
 
 | | OXA-48 carbapanemase, doripenem complex | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P99
 
 | | OXA-48 carbapanemase, ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P97
 
 | | OXA-48 carbapanemase, imipenem complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P98
 
 | | OXA-48 carbapanemase, meropenem complex | | Descriptor: | Beta-lactamase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
4GOG
 
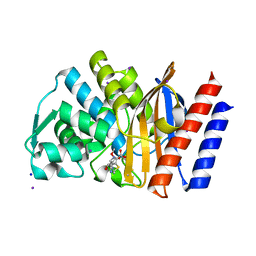 | | Crystal structure of the GES-1 imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase GES-1, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Munoz, J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
4GNU
 
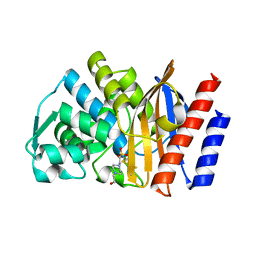 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase GES-5 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-08-17 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
4H8R
 
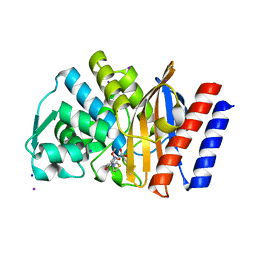 | | Imipenem complex of GES-5 carbapenemase | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Extended-spectrum beta-lactamase GES-5, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-09-23 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
4ORK
 
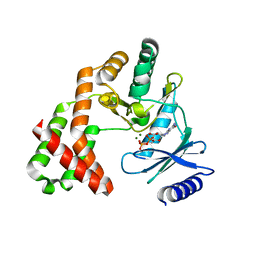 | | Crystal Structure of the Phosphotransferase Domain of the Bifunctional Aminoglycoside Resistance Enzyme AAC(6')-Ie-APH(2'')-Ia | | Descriptor: | Bifunctional AAC/APH, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Smith, C.A, Toth, M, Bhattacharya, M, Frase, H, Vakulenko, S.B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phosphotransferase domain of the bifunctional aminoglycoside-resistance enzyme AAC(6')-Ie-APH(2'')-Ia.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OH0
 
 | | Crystal structure of OXA-58 carbapenemase | | Descriptor: | Beta-lactamase OXA-58, CHLORIDE ION | | Authors: | Smith, C.A, Antunes, N.T, Toth, M, Vakulenko, S.B. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Carbapenemase OXA-58 from Acinetobacter baumannii.
Antimicrob.Agents Chemother., 58, 2014
|
|
