4I0N
 
 | | Pore forming protein | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Necrotic enteritis toxin B | | Authors: | Yan, X, Porter, C.J, Hardy, S.P, Steer, D, Smith, A.I, Quinset, N, Hughes, V, Cheung, J.K, Keyburn, A.L, Kaldhusdal, M, Moore, R.J, Bannam, T.L, Whisstock, J.C, Rood, J.I. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of the pore-forming toxin NetB from Clostridium perfringens
MBio, 4, 2013
|
|
4KKD
 
 | | The X-ray crystal structure of Mannose-binding lectin-associated serine proteinase-3 reveals the structural basis for enzyme inactivity associated with the 3MC syndrome | | Descriptor: | IMIDAZOLE, Mannan-binding lectin serine protease 1 | | Authors: | Yongqing, T, Wilmann, P.G, Reeve, S.B, Coetzer, T.H, Smith, A.I, Whisstock, J.C, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.5991 Å) | | Cite: | The X-ray Crystal Structure of Mannose-binding Lectin-associated Serine Proteinase-3 Reveals the Structural Basis for Enzyme Inactivity Associated with the Carnevale, Mingarelli, Malpuech, and Michels (3MC) Syndrome.
J.Biol.Chem., 288, 2013
|
|
2QP2
 
 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
2DUT
 
 | |
3TI9
 
 | | Crystal structure of the basic protease BprB from the ovine footrot pathogen, Dichelobacter nodosus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong, W, Whisstock, J.C, Porter, C.J. | | Deposit date: | 2011-08-20 | | Release date: | 2011-10-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S1 Pocket of a Bacterially Derived Subtilisin-like Protease Underpins Effective Tissue Destruction.
J.Biol.Chem., 286, 2011
|
|
3TI7
 
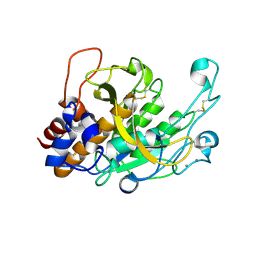 | |
2OKK
 
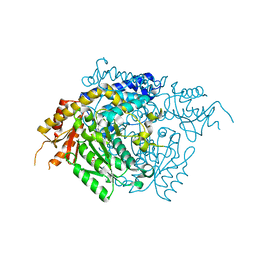 | | The X-ray crystal structure of the 65kDa isoform of Glutamic Acid Decarboxylase (GAD65) | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, Glutamate decarboxylase 2 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OKJ
 
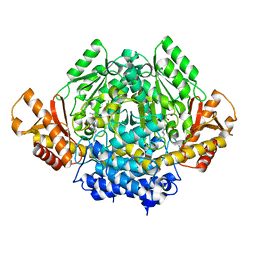 | | The X-ray crystal structure of the 67kDa isoform of Glutamic Acid Decarboxylase (GAD67) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase 1 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2PEE
 
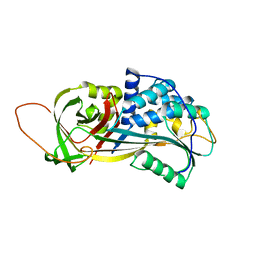 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2PEF
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Latent State | | Descriptor: | Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
3LPC
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | ACETATE ION, AprB2, CALCIUM ION, ... | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LPA
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LPD
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
4DUR
 
 | | The X-ray Crystal Structure of Full-Length type II Human Plasminogen | | Descriptor: | ACETATE ION, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Law, R.H.P, Caradoc-Davies, T, Whisstock, J.C. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The X-ray crystal structure of full-length human plasminogen
Cell Rep, 1, 2012
|
|
4DUU
 
 | |
2H4R
 
 | |
2H4Q
 
 | |
2H4P
 
 | |
