5LEW
 
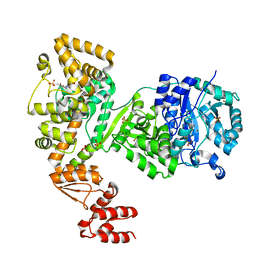 | | DNA polymerase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA polymerase III subunit alpha, SULFATE ION, ... | | Authors: | Banos-Mateos, S, Lang, U.F, Maslen, S.L, Skehel, J.M, Lamers, M.H. | | Deposit date: | 2016-06-30 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-fidelity DNA replication in Mycobacterium tuberculosis relies on a trinuclear zinc center.
Nat Commun, 8, 2017
|
|
6GLC
 
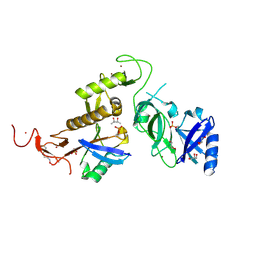 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
7OLE
 
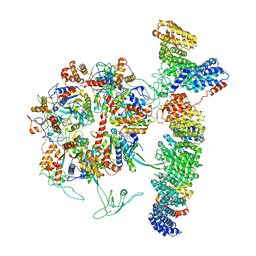 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
5NBB
 
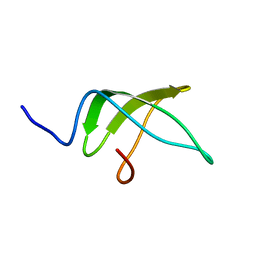 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NB9
 
 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
4PSI
 
 | | PIH1D1/phospho-Tel2 complex | | Descriptor: | PIH1 domain-containing protein 1, Telomere length regulation protein TEL2 homolog | | Authors: | Smerdon, S.J, Boulton, S.J, Stach, L, Flower, T.G, Horejsi, Z. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Phosphorylation-Dependent PIH1D1 Interactions Define Substrate Specificity of the R2TP Cochaperone Complex.
Cell Rep, 7, 2014
|
|
4PSF
 
 | | PIH1D1 N-terminal domain | | Descriptor: | PIH1 domain-containing protein 1 | | Authors: | Smerdon, S.J, Boulton, S.J, Stach, L, Flower, T.G, Horejsi, Z. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Phosphorylation-Dependent PIH1D1 Interactions Define Substrate Specificity of the R2TP Cochaperone Complex.
Cell Rep, 7, 2014
|
|
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR3
 
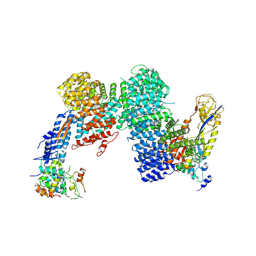 | | CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR2
 
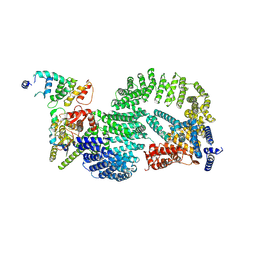 | | CAND1-CUL1-RBX1-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR4
 
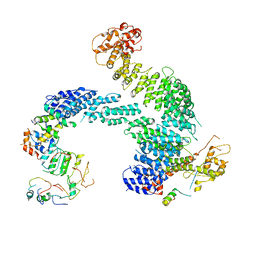 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
7ZGR
 
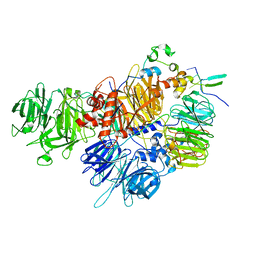 | |
7ZGQ
 
 | |
7ZGP
 
 | |
6G76
 
 | |
6G78
 
 | | RSK4 N-terminal Kinase Domain S232E in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6 | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
6G77
 
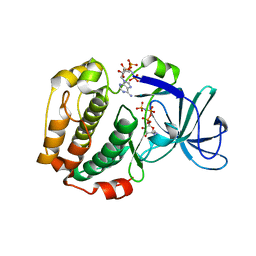 | | RSK4 N-terminal Kinase Domain in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6, ZINC ION | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
5JIG
 
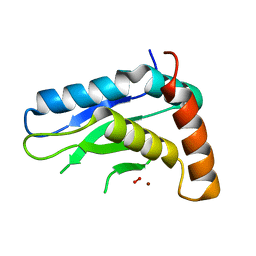 | | Crytsal structure of Wss1 from S. pombe | | Descriptor: | NICKEL (II) ION, OXYGEN MOLECULE, Ubiquitin and WLM domain-containing metalloprotease SPCC1442.07c | | Authors: | Groll, M, Stingele, J, Boulton, S. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-09 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN.
Mol.Cell, 64, 2016
|
|
5LN5
 
 | |
6YBT
 
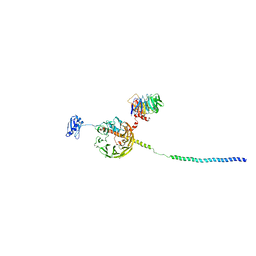 | | Structure of a human 48S translational initiation complex - eIF3bgi | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBV
 
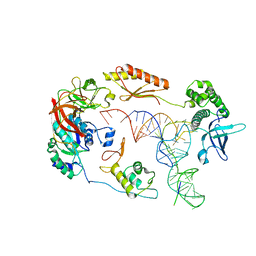 | | Structure of a human 48S translational initiation complex - eIF2-TC | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBS
 
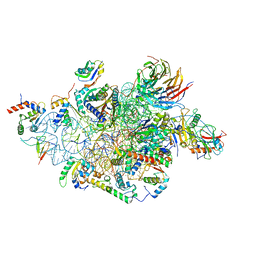 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBW
 
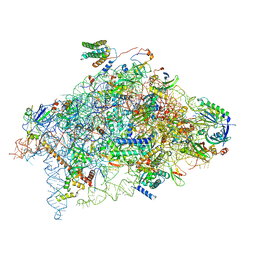 | | Structure of a human 48S translational initiation complex - 40S body | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBD
 
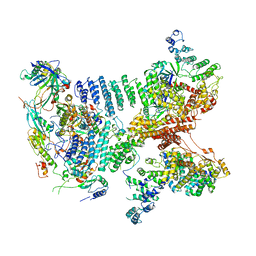 | | Structure of a human 48S translational initiation complex - eIF3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14, 40S ribosomal protein S17, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
