1GUH
 
 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | Authors: | Sinning, I, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1993-02-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
1SUR
 
 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | Descriptor: | PAPS REDUCTASE | | Authors: | Sinning, I, Savage, H. | | Deposit date: | 1998-04-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
4WJS
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODU
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CR2
 
 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CQZ
 
 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4WFM
 
 | | Structure of the complete bacterial SRP Alu domain | | Descriptor: | Bacillus subtilis small cytoplasmic RNA (scRNA),RNA, COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Kempf, G, Wild, K, Sinning, I. | | Deposit date: | 2014-09-15 | | Release date: | 2014-10-15 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the complete bacterial SRP Alu domain.
Nucleic Acids Res., 42, 2014
|
|
4WFL
 
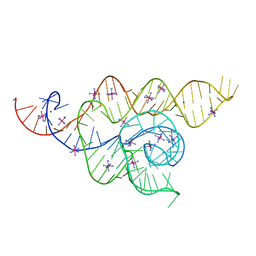 | |
1Y10
 
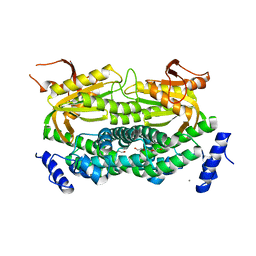 | | Mycobacterial adenylyl cyclase Rv1264, holoenzyme, inhibited state | | Descriptor: | CALCIUM ION, Hypothetical protein Rv1264/MT1302, PENTAETHYLENE GLYCOL | | Authors: | Tews, I, Findeisen, F, Sinning, I, Schultz, A, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme
Science, 308, 2005
|
|
1Y11
 
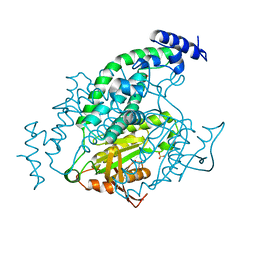 | | Mycobacterial adenylyl cyclase Rv1264, holoenzyme, active state | | Descriptor: | GLYCEROL, Hypothetical protein Rv1264/MT1302, PENTAETHYLENE GLYCOL, ... | | Authors: | Tews, I, Findeisen, F, Sinning, I, Schultz, A, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme
Science, 308, 2005
|
|
4UE4
 
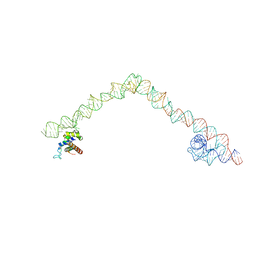 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UE5
 
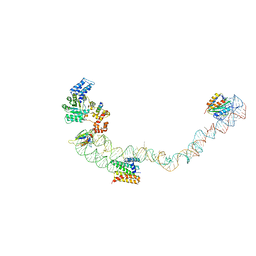 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8ONX
 
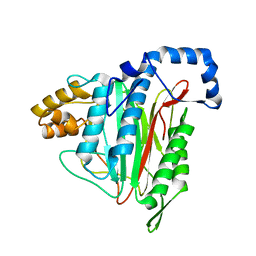 | | High resolution structure of Chaetomium thermophilum MAP2 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONY
 
 | | Human Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONZ
 
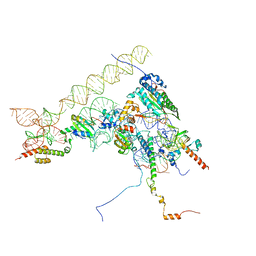 | | Chaetomium thermophilum Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L25-like protein, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8OO0
 
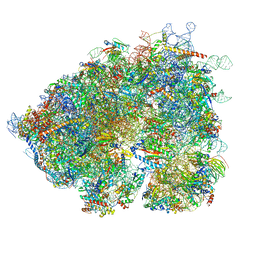 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
3BB1
 
 | | Crystal structure of Toc34 from Pisum sativum in complex with Mg2+ and GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
7OJU
 
 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Weidenhausen, J, Kopp, J, Sinning, I. | | Deposit date: | 2021-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
7Q72
 
 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
3B9Q
 
 | | The crystal structure of cpFtsY from Arabidopsis thaliana | | Descriptor: | Chloroplast SRP receptor homolog, alpha subunit CPFTSY, MALONATE ION | | Authors: | Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site
Febs Lett., 581, 2007
|
|
