1USZ
 
 | | SeMet AfaE-3 adhesin from Escherichia Coli | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1USQ
 
 | | Complex of E. Coli DraE adhesin with Chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, ... | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-11-27 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1UT2
 
 | | AfaE-3 adhesin from Escherichia Coli | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1UT1
 
 | | DraE adhesin from Escherichia Coli | | Descriptor: | 1,2-ETHANEDIOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
1RXL
 
 | | Solution structure of the engineered protein Afae-dsc | | Descriptor: | Afimbrial adhesin AFA-III | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, du Merle, L, Barlow, P.N, Medof, M.E, Smith, R.A, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-18 | | Release date: | 2005-01-11 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | An atomic resolution model for assembly, architecture, and function of the Dr adhesins.
Mol.Cell, 15, 2004
|
|
1VAZ
 
 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
1V92
 
 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
2YPL
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | AGA T-CELL RECEPTOR ALPHA CHAIN, AGA T-CELL RECEPTOR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, van der Merwe, P.A, Easterbrook, P, McMichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YPK
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-57 ALPHA CHAIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, Van Der Merwe, P.A, Easterbrook, P, Mcmichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
2BN8
 
 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | Descriptor: | CELL DIVISION ACTIVATOR CEDA | | Authors: | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | Deposit date: | 2005-03-22 | | Release date: | 2006-12-21 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
2FVN
 
 | |
2WNM
 
 | | Solution structure of Gp2 | | Descriptor: | GENE 2 | | Authors: | Camara, B, Liu, M, Shadrinc, A, Liu, B, Simpson, P, Weinzierl, R, Severinovc, K, Cota, E, Matthews, S, Wigneshweraraj, S.R. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | T7 Phage Protein Gp2 Inhibits the Escherichia Coli RNA Polymerase by Antagonizing Stable DNA Strand Separation Near the Transcription Start Site.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2X43
 
 | | STRUCTURAL BASIS OF MOLECULAR RECOGNITION BY SHERP AT MEMBRANE SURFACES | | Descriptor: | SHERP | | Authors: | Moore, B, Miles, A.J, Guerra, C.G, Simpson, P, Iwata, M, Wallace, B.A, Matthews, S.J, Smith, D.F, Brown, K.A. | | Deposit date: | 2010-02-09 | | Release date: | 2010-11-24 | | Last modified: | 2017-04-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
J.Biol.Chem., 286, 2011
|
|
4A5V
 
 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
4CB8
 
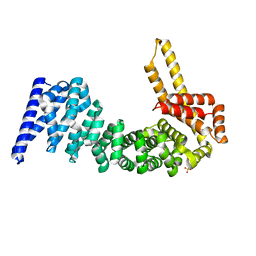 | | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha | | Descriptor: | BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Mutational Analysis Reveals that Ctnnbl1 Binds Nlss in a Manner Distinct from that of its Closest Armadillo-Relative, Karyopherin Alpha
FEBS Lett., 588, 2014
|
|
4CB9
 
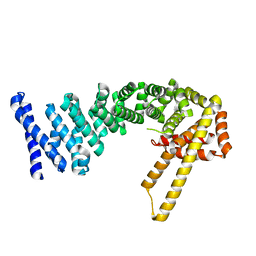 | | Structure of full-length CTNNBL1 in P43212 space group | | Descriptor: | BETA-CATENIN-LIKE PROTEIN 1 | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mutational Analysis Reveals that Ctnnbl1 Binds Nlss in a Manner Distinct from that of its Closest Armadillo-Relative, Karyopherin Alpha
FEBS Lett., 588, 2014
|
|
4CBA
 
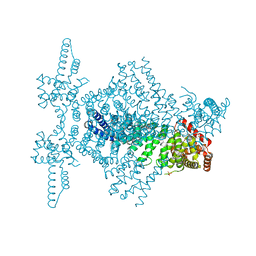 | | Structural of delta 1-76 CTNNBL1 in space group I222 | | Descriptor: | 1,2-ETHANEDIOL, BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha.
Febs Lett., 588, 2014
|
|
2IXQ
 
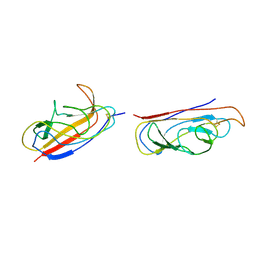 | | The solution structure of the invasive tip complex from Afa-Dr fibrils | | Descriptor: | Afimbrial adhesin AFA-III, Protein AfaD | | Authors: | Cota, E, Jones, C, Simpson, P, Altroff, H, Anderson, K.L, du Merle, L, Guignot, J, Servin, A, Le Bouguenec, C, Mardon, H, Matthews, S. | | Deposit date: | 2006-07-10 | | Release date: | 2006-09-20 | | Last modified: | 2018-12-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the invasive tip complex from Afa/Dr fibrils.
Mol. Microbiol., 62, 2006
|
|
2J76
 
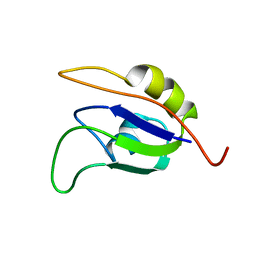 | | Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4B | | Authors: | Fleming, K, Ghuman, J, Yuan, X.M, Simpson, P, Szendroi, A, Matthews, S, Curry, S. | | Deposit date: | 2006-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2017-04-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and RNA Interactions of the RNA Recognition Motif from Eukaryotic Translation Initiation Factor 4B.
Biochemistry, 42, 2003
|
|
2JHD
 
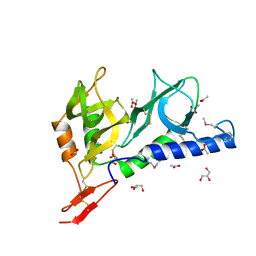 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
2JH7
 
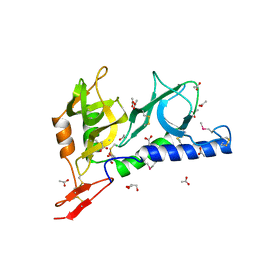 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 6'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
2JH1
 
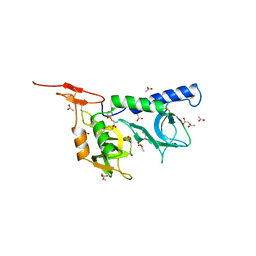 | | Crystal structure of Toxoplasma gondii micronemal protein 1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-22 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
2K2T
 
 | | Epidermal growth Factor-like domain 2 from Toxoplasma gondii Microneme protein 6 | | Descriptor: | Micronemal protein 6 | | Authors: | Sawmynaden, K, Saouros, S, Marchant, J, Simpson, P, Matthews, S. | | Deposit date: | 2008-04-11 | | Release date: | 2009-02-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural insights into microneme protein assembly reveal a new mode of EGF domain recognition.
Embo Rep., 9, 2008
|
|
2KUB
 
 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
