1B3Q
 
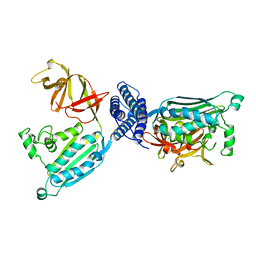 | | CRYSTAL STRUCTURE OF CHEA-289, A SIGNAL TRANSDUCING HISTIDINE KINASE | | Descriptor: | MERCURY (II) ION, PROTEIN (CHEMOTAXIS PROTEIN CHEA) | | Authors: | Bilwes, A.M, Alex, L.A, Crane, B.R, Simon, M.I. | | Deposit date: | 1998-12-14 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CheA, a signal-transducing histidine kinase.
Cell(Cambridge,Mass.), 96, 1999
|
|
2ASR
 
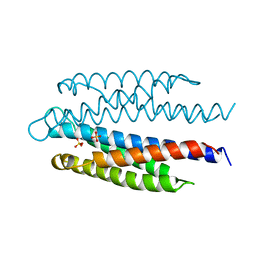 | |
1I5B
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE | | Descriptor: | ACETATE ION, CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5C
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1TQG
 
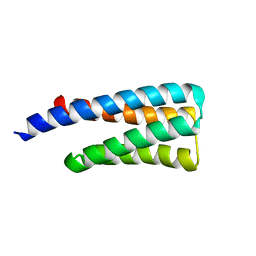 | | CheA phosphotransferase domain from Thermotoga maritima | | Descriptor: | Chemotaxis protein cheA | | Authors: | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
1I59
 
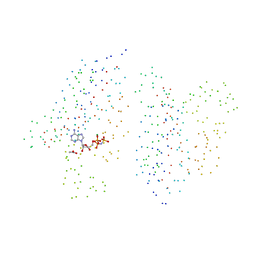 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ADPNP AND MAGENSIUM | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I58
 
 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ATP ANALOG ADPCP AND MAGNESIUM | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5A
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPCP AND MANGANESE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5D
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH TNP-ATP | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, SULFATE ION | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1U0S
 
 | | Chemotaxis kinase CheA P2 domain in complex with response regulator CheY from the thermophile thermotoga maritima | | Descriptor: | Chemotaxis protein cheA, Chemotaxis protein cheY | | Authors: | Park, S.Y, Beel, B.D, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1K0S
 
 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
3C7L
 
 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | Regulator of G-protein signaling 16 | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C7K
 
 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(o) subunit alpha, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
1EHC
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Jiang, M, Bourret, R, Simon, M, Volz, K. | | Deposit date: | 1996-03-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY.
J.Biol.Chem., 272, 1997
|
|
4TMY
 
 | | CHEY FROM THERMOTOGA MARITIMA (MG-IV) | | Descriptor: | CHEY PROTEIN, MAGNESIUM ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1C4W
 
 | | 1.9 A STRUCTURE OF A-THIOPHOSPHONATE MODIFIED CHEY D57C | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Halkides, C.J, McEvoy, M.M, Matsumura, P, Volz, K, Dahlquist, F.W. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY.
Biochemistry, 39, 2000
|
|
3TMY
 
 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1TMY
 
 | |
2TMY
 
 | |
