5FM7
 
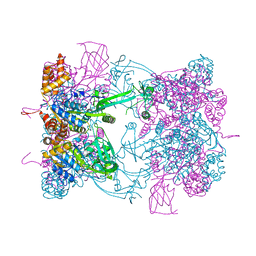 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RVB1, RVB2 | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
5FM6
 
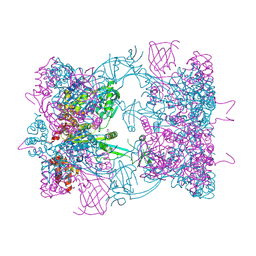 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(apo) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RVB1, ... | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
3ZHG
 
 | |
4C9F
 
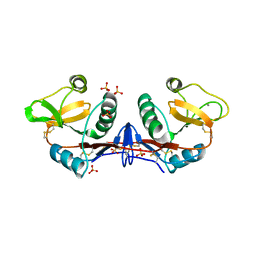 | | Structure of SIGN-R1 in complex with Sulfodextran | | Descriptor: | 4-O-sulfo-alpha-D-glucopyranose, CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Rodriguez, A, Ramirez, E, Chacon, P, Anthony, R.M, Park, C.G, Hermoso, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4CDH
 
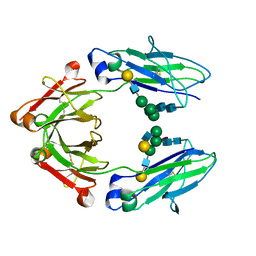 | | Crystallographic structure of the Human Igg1 alpha 2-6 sialilated Fc-Fragment | | Descriptor: | IG GAMMA-1 CHAIN C REGION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-11-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4CAJ
 
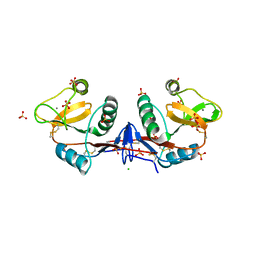 | | Crystallographic structure of the mouse SIGN-R1 CRD domain in complex with sialic acid | | Descriptor: | CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, CHLORIDE ION, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
2X8P
 
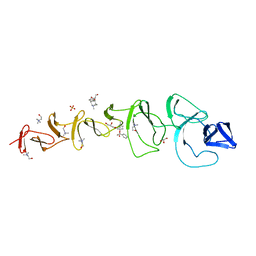 | | Crystal Structure of CbpF in Complex with Atropine by Co- Crystallization | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CHOLINE ION, CHOLINE-BINDING PROTEIN F, ... | | Authors: | Silva-Martin, N, Hermoso, J.A. | | Deposit date: | 2010-03-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of Cbpf Complexed with Atropine and Ipratropium Reveal Clues for the Design of Novel Antimicrobials Against Streptococcus Pneumoniae.
Biochim.Biophys.Acta, 1840, 2013
|
|
2X8O
 
 | |
4CVD
 
 | |
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2C
 
 | | crystal structure of AmpD Apoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2X8M
 
 | |
