6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1LVG
 
 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
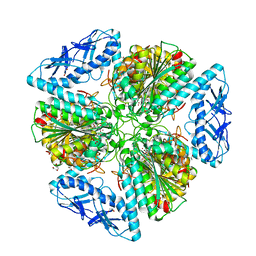 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
5T1Q
 
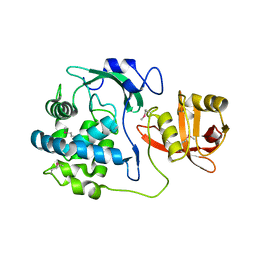 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
5SV5
 
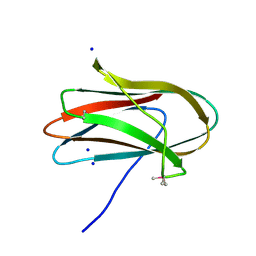 | | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis. | | Descriptor: | Microbial collagenase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.
To Be Published
|
|
3DZC
 
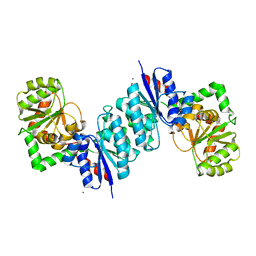 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
6DVV
 
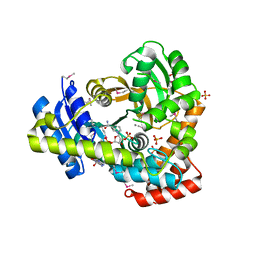 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DXN
 
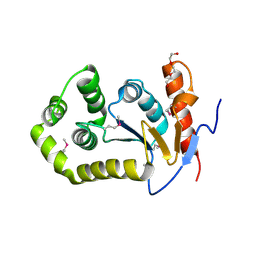 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | Descriptor: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6E5Y
 
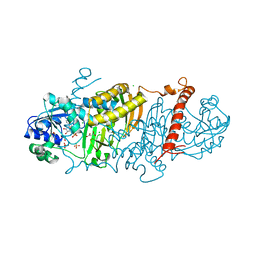 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
6E85
 
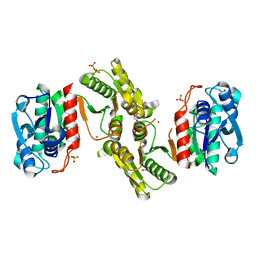 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
2AUA
 
 | | Structure of BC2332: A Protein of Unknown Function from Bacillus cereus | | Descriptor: | hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-27 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of BC2332: A hypothetical protein from Bacillus cereus
To be Published
|
|
2AP1
 
 | | Crystal structure of the putative regulatory protein | | Descriptor: | SODIUM ION, ZINC ION, putative regulator protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative regulatory protein
To be Published
|
|
2BAS
 
 | | Crystal Structure of the Bacillus subtilis YkuI Protein, with an EAL Domain. | | Descriptor: | BETA-MERCAPTOETHANOL, YkuI protein | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Miller, D.J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains.
J.Biol.Chem., 284, 2009
|
|
6MUQ
 
 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
6MUK
 
 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
6N7F
 
 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
2FOR
 
 | | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate | | Descriptor: | Geranyltranstransferase, ISOPENTYL PYROPHOSPHATE, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate
To be Published
|
|
2FZV
 
 | | Crystal Structure of an apo form of a Flavin-binding Protein from Shigella flexneri | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative arsenical resistance protein | | Authors: | Vorontsov, I.I, Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity
Protein Sci., 16, 2007
|
|
7R6S
 
 | | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia | | Descriptor: | Putative bacteriophage protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Brunzelle, J.S, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia
To Be Published
|
|
7RJ3
 
 | | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycogen accumulation regulator GarA | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis.
To Be Published
|
|
