3VTE
 
 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
3W2E
 
 | | Crystal structure of oxidation intermediate (20 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2F
 
 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2G
 
 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2H
 
 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2I
 
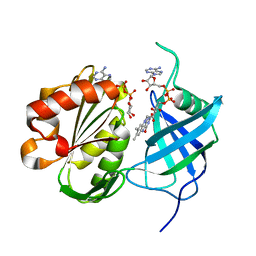 | | Crystal structure of re-oxidized form (60 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
2ZIL
 
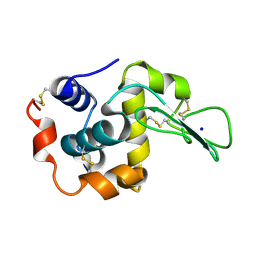 | | Crystal Structure of Human Lysozyme from Urine | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Tamada, T, Kuroki, R, Koshiba, T. | | Deposit date: | 2008-02-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preparation and characterization of methyonyl-lysine attached human lysozyme expressed in Escherichia coli and its effective conversion to the authentic-like protein
To be Published
|
|
2ZIK
 
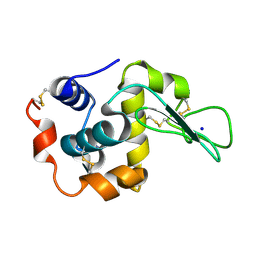 | |
2ZIJ
 
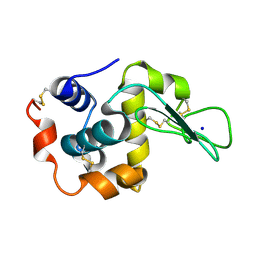 | |
3FX5
 
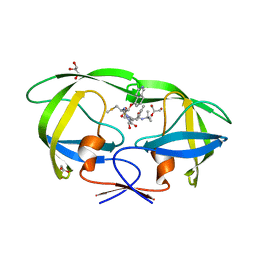 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2021-11-10 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3W5H
 
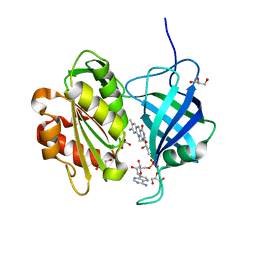 | | Ultra-high resolution structure of NADH-cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADH-cytochrome b5 reductase 3 | | Authors: | Takeda, K, Ohno, H, Kosugi, M, Takaba, K, Miki, K. | | Deposit date: | 2013-01-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
