6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KMF
 
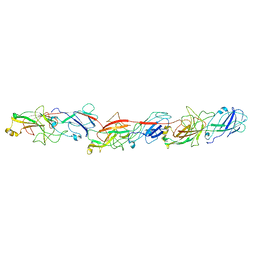 | | FimA type V pilus from P.gingivalis | | Descriptor: | Major fimbrium subunit FimA type-1 | | Authors: | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JZJ
 
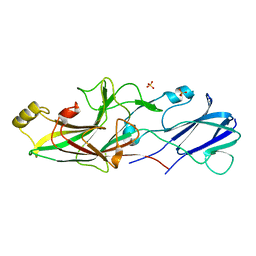 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JZK
 
 | | Structure of FimA type-1 (FimA1) prepilin of the type V major fimbrium | | Descriptor: | GLYCEROL, Major fimbrium subunit FimA type-1, S,R MESO-TARTARIC ACID, ... | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6KJK
 
 | | Structure of the N-terminal domain of PorA | | Descriptor: | N-terminal domain of PorA (28-171) | | Authors: | Handa, Y, Sato, K, Shoji, M, Nakayama, K, Imada, K. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PorA, a conserved C-terminal domain-containing protein, impacts the PorXY-SigP signaling of the type IX secretion system.
Sci Rep, 10, 2020
|
|
7WIS
 
 | |
7WIR
 
 | |
3LIU
 
 | |
3PAY
 
 | |
3R4R
 
 | |
4DGU
 
 | |
4EPS
 
 | |
8J6G
 
 | |
3GF8
 
 | |
4QDG
 
 | |
4Q98
 
 | |
4QB7
 
 | |
4RDB
 
 | |
4H40
 
 | |
4GPV
 
 | |
4JG5
 
 | |
4JRF
 
 | |
4K4K
 
 | |
5CAG
 
 | |
5Y1A
 
 | | HBP35 of Porphyromonas gingivalis | | Descriptor: | 35 kDa hemin binding protein | | Authors: | Kakuda, S, Suzuki, M, Sato, K. | | Deposit date: | 2017-07-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system.
Mol. Microbiol., 110, 2018
|
|
