1CH4
 
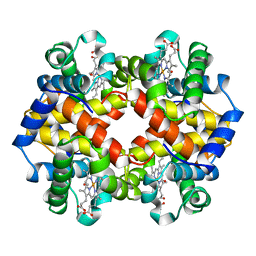 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | Descriptor: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | Deposit date: | 1998-06-11 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
1C1F
 
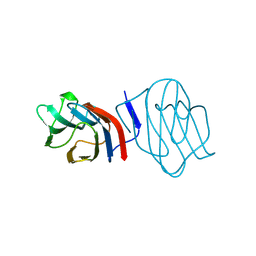 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1C1L
 
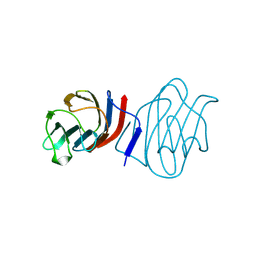 | | LACTOSE-LIGANDED CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1G01
 
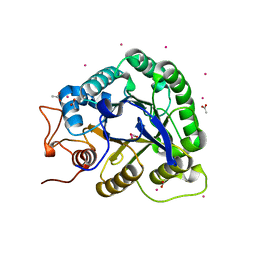 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G0C
 
 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1IS4
 
 | | LACTOSE-LIGANDED CONGERIN II | | Descriptor: | CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Conger Eel Galectin (Congerin II) at 1.45 A Resolution: Implication for the Accelerated Evolution of a New Ligand-Binding Site Following Gene Duplication
J.Mol.Biol., 321, 2002
|
|
1IS3
 
 | | LACTOSE AND MES-LIGANDED CONGERIN II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS5
 
 | | Ligand free Congerin II | | Descriptor: | Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS6
 
 | | MES-Liganded Congerin II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1WSD
 
 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
2DIE
 
 | | Alkaline alpha-amylase AmyK from Bacillus sp. KSM-1378 | | Descriptor: | CALCIUM ION, SODIUM ION, amylase | | Authors: | Shirai, T, Igarashi, K, Ozawa, T, Hagihara, H, Kobayashi, T, Ozaki, K, Ito, S. | | Deposit date: | 2006-03-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ancestral sequence evolutionary trace and crystal structure analyses of alkaline alpha-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins
Proteins, 66, 2007
|
|
2ZE0
 
 | | Alpha-glucosidase GSJ | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Shirai, T, Hung, V.S, Morinaka, K, Kobayashi, T, Ito, S. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GH13 alpha-glucosidase GSJ from one of the deepest sea bacteria
Proteins, 73, 2008
|
|
2ZX0
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX4
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX3
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
4KMI
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with PO4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | Authors: | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | Deposit date: | 2001-09-05 | | Release date: | 2003-11-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
7DDR
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDS
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative | | Descriptor: | Ancestral myoglobin aMbSp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDT
 
 | | Ancestral myoglobin aMbSe of Enaliarctos relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSe, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDU
 
 | | Elephant seal myoglobin esMb | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
1WLC
 
 | | Congerin II Y16S/T88I double mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shionyu-Mitsuyama, C, Ito, Y, Konno, A, Miwa, Y, Ogawa, T, Muramoto, K, Shirai, T. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro evolutionary thermostabilization of congerin II: a limited reproduction of natural protein evolution by artificial selection pressure
J.Mol.Biol., 347, 2005
|
|
1WLD
 
 | | Congerin II T88I single mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shionyu-Mitsuyama, C, Ito, Y, Konno, A, Miwa, Y, Ogawa, T, Muramoto, K, Shirai, T. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In vitro evolutionary thermostabilization of congerin II: a limited reproduction of natural protein evolution by artificial selection pressure
J.Mol.Biol., 347, 2005
|
|
