1J03
 
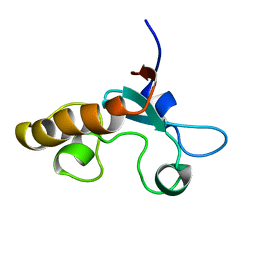 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
1VEE
 
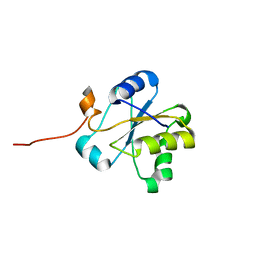 | | NMR structure of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana | | Descriptor: | proline-rich protein family | | Authors: | Pantoja-Uceda, D, Lopez-Mendez, B, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana
Protein Sci., 14, 2005
|
|
1VDY
 
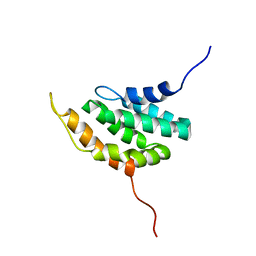 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
1UHT
 
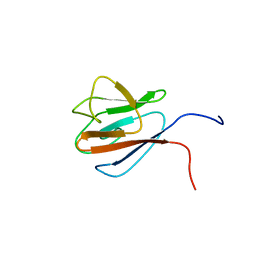 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | Descriptor: | expressed protein | | Authors: | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
3JRS
 
 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
1CQZ
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2DCQ
 
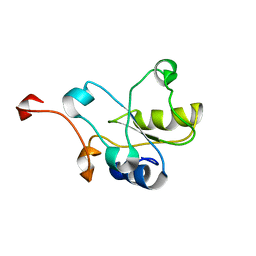 | |
1UL5
 
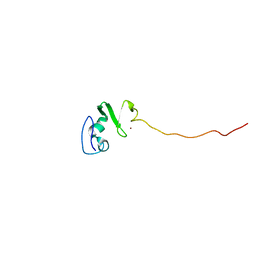 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 7 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1UL4
 
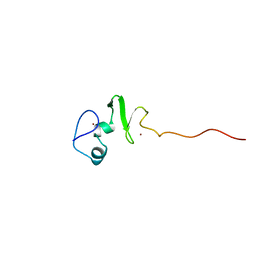 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 4 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1WJ0
 
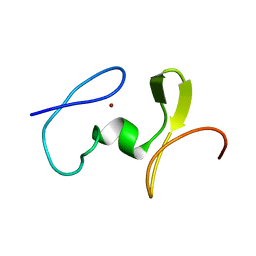 | |
1WIJ
 
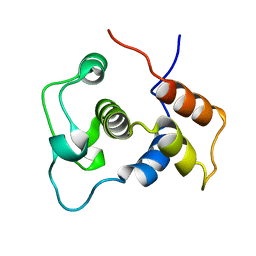 | | Solution Structure of the DNA-Binding Domain of Ethylene-Insensitive3-Like3 | | Descriptor: | ETHYLENE-INSENSITIVE3-like 3 protein | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3.
J.Mol.Biol., 348, 2005
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | Descriptor: | Probable WRKY transcription factor 4, ZINC ION | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
1WID
 
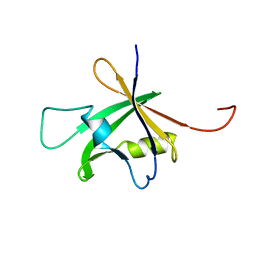 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
2DCP
 
 | |
