1H6L
 
 | |
1O9N
 
 | |
1OBI
 
 | |
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1O9O
 
 | |
1O9Q
 
 | |
1OCK
 
 | |
1OCH
 
 | |
1OBJ
 
 | |
1O9P
 
 | |
1OCL
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
 | |
1CVM
 
 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1GS3
 
 | |
1QLG
 
 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
1V0D
 
 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
5IB0
 
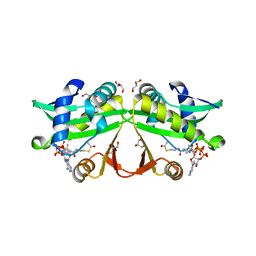 | | PA4534: acetyl CoA complex | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Uncharacterized protein PA4534 | | Authors: | Choe, J, Shin, S. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of PA4534
To Be Published
|
|
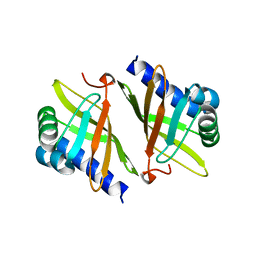
1OCV
 
 | |
5Z8T
 
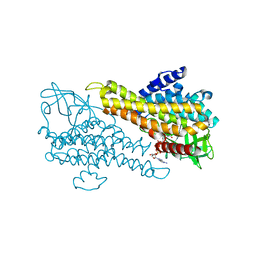 | | Flavin-containing monooxygenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choe, J, Moon, H, Shin, S. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of putative Flavin-dependent monooxgenase
To Be Published
|
|
4Y5R
 
 | | Crystal Structure of a T67A MauG/pre-Methylamine Dehydrogenase Complex | | Descriptor: | CALCIUM ION, HEME C, Methylamine dehydrogenase heavy chain, ... | | Authors: | Li, C, Wilmot, C.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Biochim.Biophys.Acta, 1847, 2015
|
|
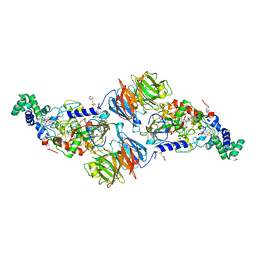
4O1Q
 
 | |
2X29
 
 | | Crystal structure of human4-1BB ligand ectodomain | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 9 | | Authors: | Won, E.Y, Cho, H.S. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the Trimer of Human 4-1Bb Ligand is Unique Among Members of the Tumor Necrosis Factor Superfamily.
J.Biol.Chem., 285, 2010
|
|
5KTG
 
 | |
3QID
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
