1DIO
 
 | | DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | COBALAMIN, POTASSIUM ION, PROTEIN (DIOL DEHYDRATASE), ... | | Authors: | Shibata, N, Masuda, J, Tobimatsu, T, Toraya, T, Suto, K, Morimoto, Y, Yasuoka, N. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase.
Structure Fold.Des., 7, 1999
|
|
1A7V
 
 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS PALUSTRIS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shibata, N, Iba, S, Misaki, S, Meyer, T.E, Bartsch, R.G, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1998-03-18 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Basis for monomer stabilization in Rhodopseudomonas palustris cytochrome c' derived from the crystal structure.
J.Mol.Biol., 284, 1998
|
|
1IWB
 
 | | Crystal structure of diol dehydratase | | Descriptor: | COBALAMIN, DIOL DEHYDRATASE alpha chain, DIOL DEHYDRATASE beta chain, ... | | Authors: | Shibata, N, Masuda, J, Morimoto, Y, Yasuoka, N, Toraya, T. | | Deposit date: | 2002-05-01 | | Release date: | 2003-05-01 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-induced conformational change of a coenzyme B12-dependent enzyme: crystal structure of the substrate-free form of diol dehydratase
Biochemistry, 41, 2002
|
|
1IUZ
 
 | | PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, SULFATE ION | | Authors: | Shibata, N. | | Deposit date: | 1996-10-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel insight into the copper-ligand geometry in the crystal structure of Ulva pertusa plastocyanin at 1.6-A resolution. Structural basis for regulation of the copper site by residue 88.
J.Biol.Chem., 274, 1999
|
|
1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
7XRL
 
 | | Diol dehydratase complexed with AdoMeCbl and 1,2-propanediol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRK
 
 | | Diol dehydratase complexed with AdoMeCbl | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, CALCIUM ION, COBALAMIN, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRM
 
 | | Ethanolamine ammonia-lyase complexed with AdoMeCbl | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, COBALAMIN, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRN
 
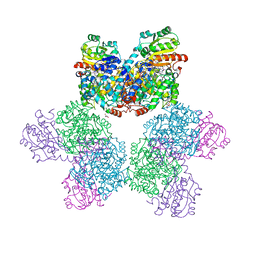 | | Ethanolamine ammonia-lyase complexed with AdoMeCbl in the presence of substrate | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, COBALAMIN, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
2D0O
 
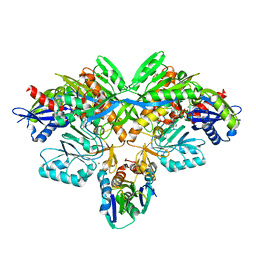 | | Structure of diol dehydratase-reactivating factor complexed with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
2D0P
 
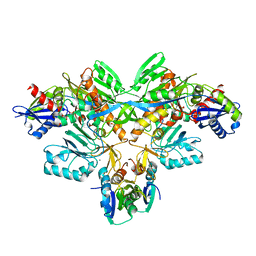 | | Structure of diol dehydratase-reactivating factor in nucleotide free form | | Descriptor: | CALCIUM ION, SULFATE ION, diol dehydratase-reactivating factor large subunit, ... | | Authors: | Shibata, N, Mori, K, Hieda, N, Higuchi, Y, Yamanishi, M, Toraya, T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor
Structure, 13, 2005
|
|
5YRT
 
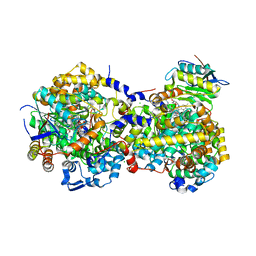 | |
5YSN
 
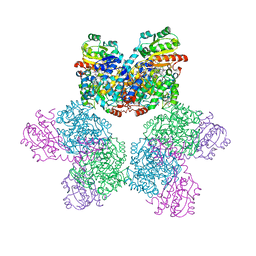 | | Ethanolamine ammonia-lyase, AdoCbl/substrate-free | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Ethanolamine ammonia-lyase heavy chain, ... | | Authors: | Shibata, N. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Direct Participation of a Peripheral Side Chain of a Corrin Ring in Coenzyme B12Catalysis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YSH
 
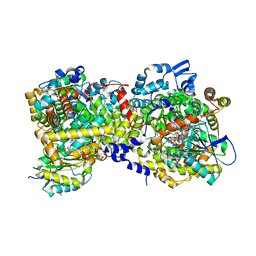 | |
5YRV
 
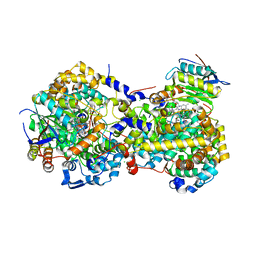 | |
5YSR
 
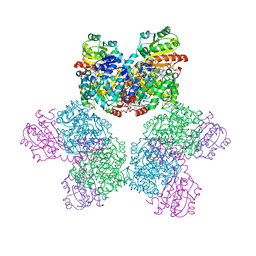 | | Ethanolamine ammonia-lyase, AdoCbl/2-amino-1-propanol | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Ethanolamine ammonia-lyase heavy chain, ... | | Authors: | Shibata, N. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct Participation of a Peripheral Side Chain of a Corrin Ring in Coenzyme B12Catalysis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1UC4
 
 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
2D5G
 
 | |
2E83
 
 | |
3ABQ
 
 | |
3ABR
 
 | |
3ABO
 
 | |
3ABS
 
 | |
1WSP
 
 | | Crystal structure of axin dix domain | | Descriptor: | Axin 1 protein, BENZOIC ACID, MERCURY (II) ION | | Authors: | Shibata, N, Hanamura, T, Yamamoto, R, Ueda, Y, Yamamoto, H, Kikuchi, A, Higuchi, Y. | | Deposit date: | 2004-11-08 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of axin dix domain
to be published
|
|
3ANY
 
 | |
