2QZS
 
 | | Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSb) | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, ADENOSINE-5'-DIPHOSPHATE, alpha-D-glucopyranose, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2007-08-17 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structures of the Open and Catalytically Competent Closed Conformation of Escherichia coli Glycogen Synthase.
J.Biol.Chem., 284, 2009
|
|
2R4U
 
 | | Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSd) | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2007-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase.
J.Biol.Chem., 284, 2009
|
|
2R4T
 
 | | Crystal Structure of Wild-type E.coli GS in Complex with ADP and Glucose(wtGSc) | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2007-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase.
J.Biol.Chem., 284, 2009
|
|
3D1J
 
 | |
3CX4
 
 | |
3COP
 
 | |
3GUH
 
 | | Crystal Structure of Wild-type E.coli GS in complex with ADP and DGM | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 1,5-anhydro-D-glucitol, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Crystal Structures of the Open and Catalytically Competent Closed Conformation of Escherichia coli Glycogen Synthase.
J.Biol.Chem., 284, 2009
|
|
5FOI
 
 | | Crystal structure of mycinamicin VIII C21 methyl hydroxylase MycCI from Micromonospora griseorubida bound to mycinamicin VIII | | Descriptor: | GLYCEROL, MYCINAMICIN VIII C21 METHYL HYDROXYLASE, Mycinamicin VIII, ... | | Authors: | Demars, M, Sheng, F, Podust, L.M, Sherman, D.H. | | Deposit date: | 2015-11-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and Structural Characterization of Mycci, a Versatile P450 Biocatalyst from the Mycinamicin Biosynthetic Pathway.
Acs Chem.Biol., 11, 2016
|
|
4A65
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with AuCN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4UP3
 
 | | Crystal structure of the mutant C140S,C286Q thioredoxin reductase from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Vieira, D.F. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
5E70
 
 | | Crystal structure of Ecoli Branching Enzyme with gamma cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E6Z
 
 | | Crystal structure of Ecoli Branching Enzyme with beta cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E6Y
 
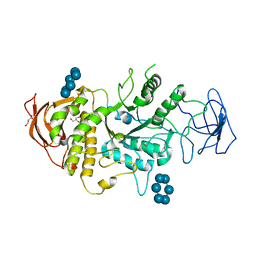 | | Crystal structure of E.Coli branching enzyme in complex with alpha cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4A5L
 
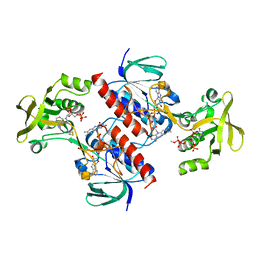 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Podust, L.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CCQ
 
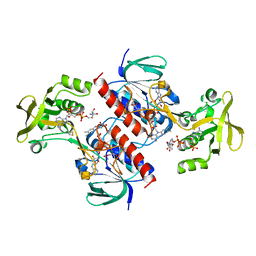 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CCR
 
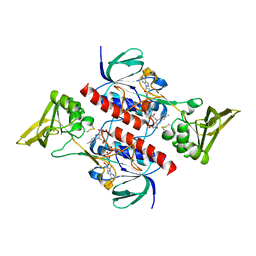 | | Crystal structure of the thioredoxin reductase apoenzyme from Entamoeba histolytica in the absence of the NADP cofactor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, THIOREDOXIN REDUCTASE | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CBQ
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with auranofin Au(I) bound to Cys286 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-16 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
3ISP
 
 | | Crystal structure of ArgP from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator Rv1985c/MT2039 | | Authors: | Zhou, X, Lou, Z, Sheng, F, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2009-08-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ArgP from Mycobacterium tuberculosis Confirms Two Distinct Conformations of Full-length LysR Transcriptional Regulators and Reveals Its Function in DNA Binding and Transcriptional Regulation.
J.Mol.Biol., 2009
|
|
