1NIZ
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1NJ0
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
6FUY
 
 | | Crystal structure of human full-length vinculin-T12-A974K (residues 1-1066) | | Descriptor: | CALCIUM ION, Vinculin | | Authors: | Chorev, D.S, Volberg, T, Livne, A, Eisenstein, M, Martins, B, Kam, Z, Jockusch, B.M, Medalia, O, Sharon, M, Geiger, B. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational states during vinculin unlocking differentially regulate focal adhesion properties.
Sci Rep, 8, 2018
|
|
1U6U
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1U6V
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
3OGM
 
 | | Structure of COI1-ASK1 in complex with coronatine and the JAZ1 degron | | Descriptor: | (1S,2S)-2-ethyl-1-({[(3aS,4S,6R,7aS)-6-ethyl-1-oxooctahydro-1H-inden-4-yl]carbonyl}amino)cyclopropanecarboxylic acid, Coronatine-insensitive protein 1, JAZ1 degron peptide, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
3OGL
 
 | | Structure of COI1-ASK1 in complex with JA-isoleucine and the JAZ1 degron | | Descriptor: | Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, N-({(1R,2S)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
3OGK
 
 | | Structure of COI1-ASK1 in complex with coronatine and an incomplete JAZ1 degron | | Descriptor: | (1S,2S)-2-ethyl-1-({[(3aS,4S,6R,7aS)-6-ethyl-1-oxooctahydro-1H-inden-4-yl]carbonyl}amino)cyclopropanecarboxylic acid, Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
6GC2
 
 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
2P1P
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1N
 
 | | Mechanism of Auxin Perception by the TIR1 Ubiqutin Ligase | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, C, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1O
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, NAPHTHALEN-1-YL-ACETIC ACID, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1M
 
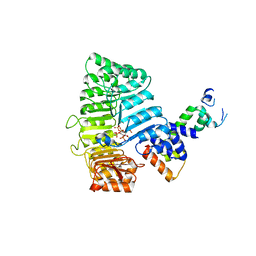 | | TIR1-ASK1 complex structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, TRANSPORT INHIBITOR RESPONSE 1 protein | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-05 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1Q
 
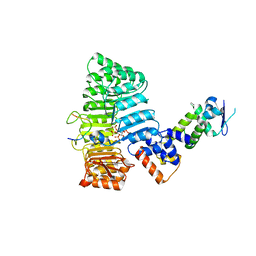 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase.
Nature, 446, 2007
|
|
2ESX
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (minimized average structure) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
2ESZ
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (ensemble) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
6TU3
 
 | | Rat 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Deshmukh, F.K, Polkinghorn, C.R, Elad, N, Sharon, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Comparative Structural Analysis of 20S Proteasome Ortholog Protein Complexes by Native Mass Spectrometry.
Acs Cent.Sci., 6, 2020
|
|
4Q1Q
 
 | |
2H6J
 
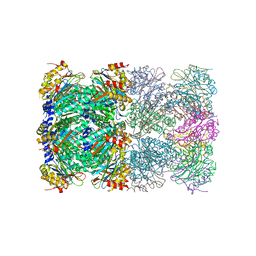 | | Crystal Structure of the Beta F145A Rhodococcus Proteasome | | Descriptor: | Proteasome alpha-type subunit 1, Proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Proteasome assembly triggers a switch required for active-site maturation.
Structure, 14, 2006
|
|
3KIH
 
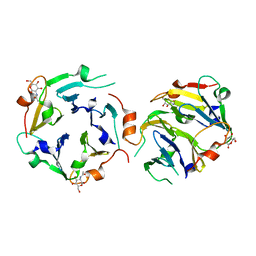 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KIF
 
 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib1-B7-18 and Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin, SULFATE ION | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4RAP
 
 | | Crystal structure of bacterial iron-containing dodecameric glycosyltransferase TibC from enterotoxigenic E.coli H10407 | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase TibC | | Authors: | Yao, Q, Lu, Q, Xu, Y, Shao, F. | | Deposit date: | 2014-09-10 | | Release date: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family.
Elife, 3, 2014
|
|
8PHL
 
 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-06-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8Q0C
 
 | | Human carbonic anhydrase II containing 3-fluorotyrosine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8P6U
 
 | | Human carbonic anhydrase II containing 5-fluorotryptophanes | | Descriptor: | BENZOIC ACID, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
