2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | Descriptor: | Bacterial Ig-like domain, group 2 | | Authors: | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
3CW3
 
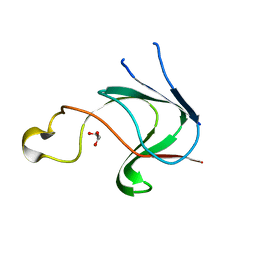 | | Crystal structure of AIM1g1 | | Descriptor: | Absent in melanoma 1 protein, GLYCEROL | | Authors: | Aravind, P, Sankaranarayanan, R, Sharma, Y. | | Deposit date: | 2008-04-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Exploring the limits of sequence and structure in a variant betagamma-crystallin domain of the protein absent in melanoma-1 (AIM1).
J.Mol.Biol., 381, 2008
|
|
2K1W
 
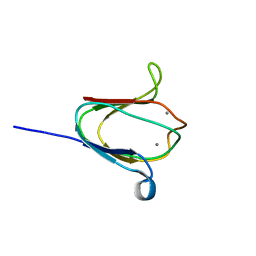 | | NMR solution structure of M-crystallin in calcium loaded form(holo). | | Descriptor: | Beta/gama crystallin family protein, CALCIUM ION | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2K1X
 
 | | NMR solution structure of M-crystallin in calcium free form (apo). | | Descriptor: | Beta/gama crystallin family protein | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2KP5
 
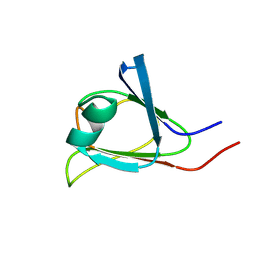 | |
5HT8
 
 | | Crystal structure of clostrillin double mutant (S17H,S19H) in complex with nickel | | Descriptor: | Beta and gamma crystallin, NICKEL (II) ION | | Authors: | Jamkhindikar, A, Srivastava, S.S, Sankaranarayanan, R. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Transition Metal-Binding, Trimeric beta gamma-Crystallin from Methane-Producing Thermophilic Archaea, Methanosaeta thermophila
Biochemistry, 56, 2017
|
|
5HT9
 
 | |
5HT7
 
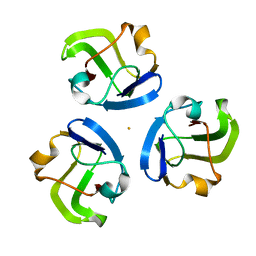 | |
4FD9
 
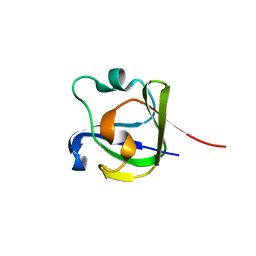 | |
3ENT
 
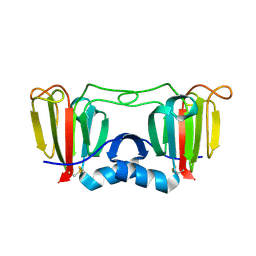 | |
3ENU
 
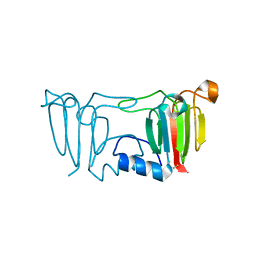 | |
3HZ2
 
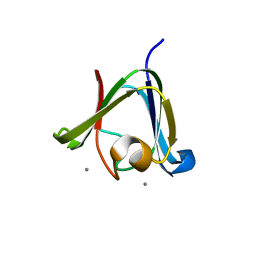 | |
3HZB
 
 | |
3I9H
 
 | |
3IAJ
 
 | |
3SNZ
 
 | |
3SO0
 
 | |
3SNY
 
 | |
3SO1
 
 | |
5Z6D
 
 | |
5Z6E
 
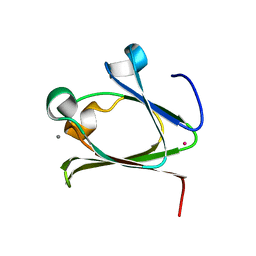 | |
7XXN
 
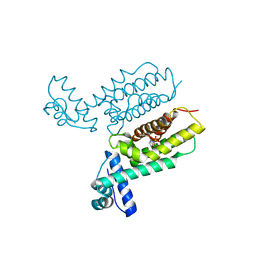 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
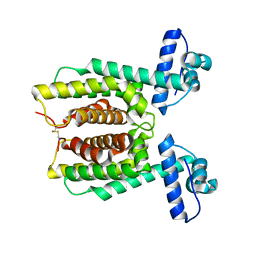 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXO
 
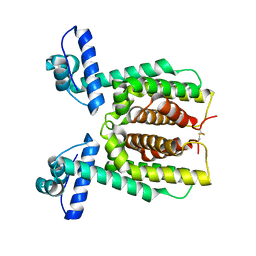 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXT
 
 | | HapR Quadruple mutant, bound to IMTVC-212 | | Descriptor: | 1-((5-phenylthiophen-2-yl)sulfonyl)-1H-pyrazole, DIMETHYL SULFOXIDE, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
