1YR2
 
 | |
1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9U
 
 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
2BKL
 
 | | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Inter-Domain Dynamics in Catalysis and Specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, N-[(BENZYLOXY)CARBONYL]-L-ALANYL-L-PROLINE, PROLYL ENDOPEPTIDASE, ... | | Authors: | Khosla, C, Shan, L, Mathews, I.I. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6WIB
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 4, ZINC ION | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W, van Dyk, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WOL
 
 | | Next generation monomeric IgG4 Fc bound to neonatal Fc receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, IgG receptor FcRn large subunit p51, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WNA
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, Immunoglobulin heavy constant gamma 4, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-22 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WMH
 
 | |
5HVW
 
 | | Monomeric IgG4 Fc | | Descriptor: | GLYCEROL, Ig gamma-4 chain C region, ZINC ION, ... | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W.F. | | Deposit date: | 2016-01-28 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation and Characterization of an IgG4 Monomeric Fc Platform.
Plos One, 11, 2016
|
|
2CGR
 
 | |
1CGS
 
 | |
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
1JVK
 
 | | THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Bourne, P.C, Ramsland, P.A, Shan, L, Fan, Z.-C, DeWitt, C.R, Shultz, B.B, Terzyan, S.S, Edmundson, A.B. | | Deposit date: | 2001-08-30 | | Release date: | 2002-05-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1NOT
 
 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3W
 
 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2FXZ
 
 | |
2FXY
 
 | |
1B2W
 
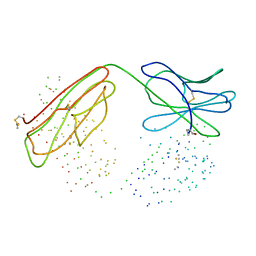 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B4J
 
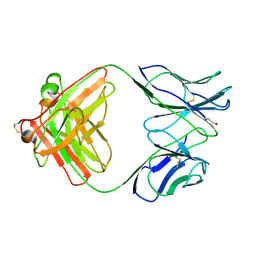 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1DQL
 
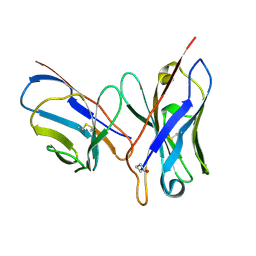 | | CRYSTAL STRUCTURE OF AN UNLIGANDED (NATIVE) FV FROM A HUMAN IGM ANTI-PEPTIDE ANTIBODY | | Descriptor: | IGM MEZ IMMUNOGLOBULIN | | Authors: | Ramsland, P.A, Shan, L, Moomaw, C.R, Slaughter, C.A, Guddat, L.W, Edmundson, A.B. | | Deposit date: | 2000-01-04 | | Release date: | 2000-10-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An unusual human IgM antibody with a protruding HCDR3 and high avidity for its peptide ligands.
Mol.Immunol., 37, 2000
|
|
2KHS
 
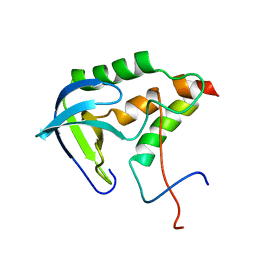 | | Solution structure of SNase121:SNase(111-143) complex | | Descriptor: | Nuclease, Thermonuclease | | Authors: | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | Deposit date: | 2009-04-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
2M00
 
 | |
