5ZK0
 
 | | Crystal structure of Peptidyl-tRNA hydrolase mutant -M71A from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Shahid, S, Kabra, A, Pal, R.K, Arora, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of methionine 71 in substrate recognition and structural integrity of bacterial peptidyl-tRNA hydrolase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6X9X
 
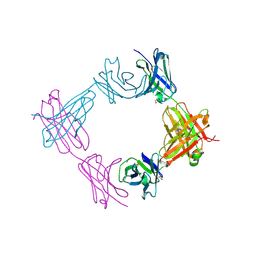 | |
5EKT
 
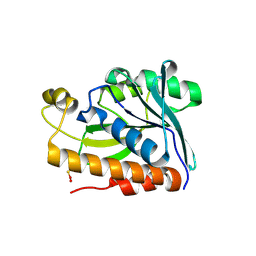 | |
5GVZ
 
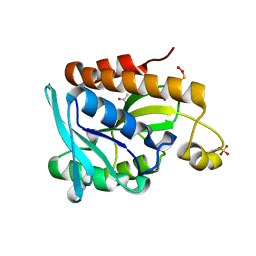 | |
5IVP
 
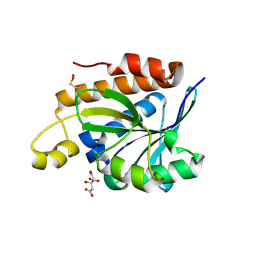 | |
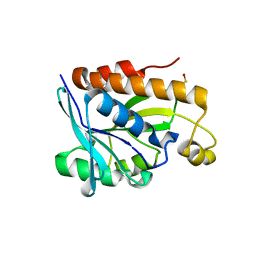
5B6J
 
 | |
4Z86
 
 | |
4ZXP
 
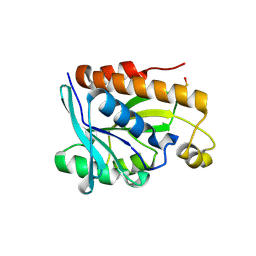 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | Descriptor: | CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
5IKE
 
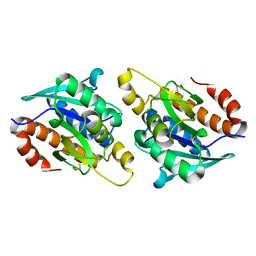 | |
5IMB
 
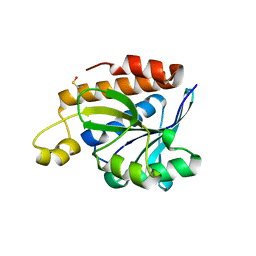 | |
8SO3
 
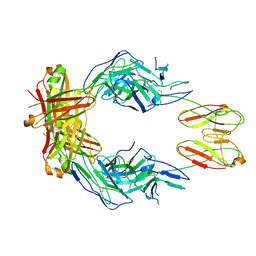 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SR0
 
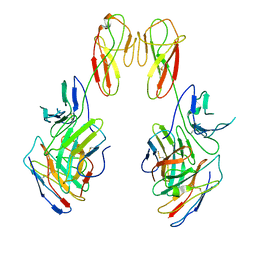 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
2MJL
 
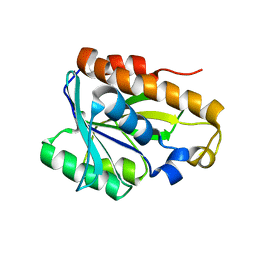 | | Solution structure of peptidyl-tRNA hyrolase from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kabra, A, Shahid, S, Yadav, R, Pulavarti, S, Shukla, V.K, Arora, A. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptidyl-tRNA hydrolase from Vibrio cholerae
To be Published
|
|
6P73
 
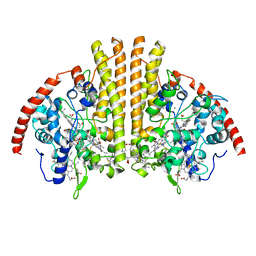 | | Cytochrome-C-nitrite reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Schmidt, M, Pacheco, A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
8FWH
 
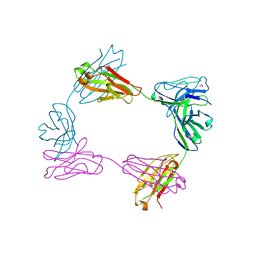 | | Crystal structure of bivalent antibody Fab fragment of Anti-human LAG3 (22D2) | | Descriptor: | 1,2-ETHANEDIOL, Anti-human LAG3 (22D2) heavy chain, Anti-human LAG3 (22D2) light chain | | Authors: | Mishra, A.K, Agnihotri, P, Mariuzza, R.A. | | Deposit date: | 2023-01-22 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
2NAF
 
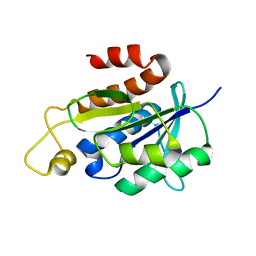 | | Solution structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Fatma, F, Kabra, A, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2015-12-23 | | Release date: | 2017-01-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of peptidyl-tRNA hydrolase from Mycobacterium smegmatis by NMR spectroscopy.
Biochim.Biophys.Acta, 1864, 2016
|
|
6WKM
 
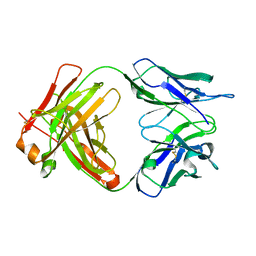 | |
7P3F
 
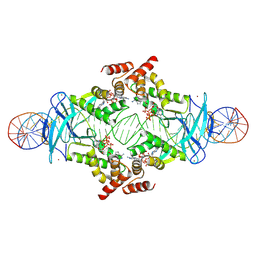 | |
7P3Q
 
 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P37
 
 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
4Z0X
 
 | |
