3RTK
 
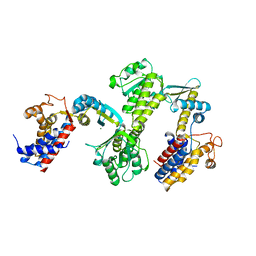 | | Crystal structure of Cpn60.2 from Mycobacterium tuberculosis at 2.8A | | Descriptor: | 60 kDa chaperonin 2, MAGNESIUM ION | | Authors: | Shahar, A, Melamed-Frank, M, Kashi, Y, Adir, N. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The dimeric structure of the Cpn60.2 chaperonin of Mycobacterium tuberculosis at 2.8 A reveals possible modes of function.
J.Mol.Biol., 412, 2011
|
|
6XXN
 
 | | Crystal structure of NB7, a nanobody targeting prostate specific membrane antigen | | Descriptor: | NB_7_a,b,c,f, NB_7_g, NB_7_h, ... | | Authors: | Shahar, A, Rosenfeld, L, Papo, N. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Nanobodies Targeting Prostate-Specific Membrane Antigen for the Imaging and Therapy of Prostate Cancer.
J.Med.Chem., 63, 2020
|
|
6HAR
 
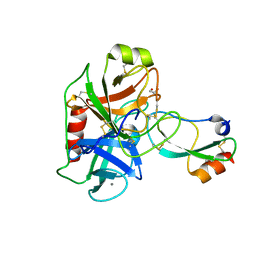 | | Crystal structure of Mesotrypsin in complex with APPI-M17C/I18F/F34C | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, CALCIUM ION, ... | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J.Biol.Chem., 294, 2019
|
|
5LXF
 
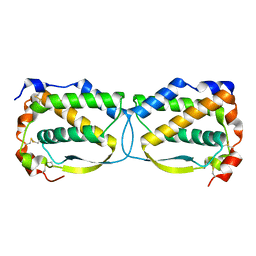 | | Crystal structure of the human Macrophage Colony Stimulating Factor M- CSF_C31S variant | | Descriptor: | Macrophage colony-stimulating factor 1 | | Authors: | Shahar, A, Papo, N, Zarivach, R, Kosloff, M, Bakhman, A, Rosenfeld, L, Zur, Y, Levaot, N. | | Deposit date: | 2016-09-21 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a monomeric variant of macrophage colony-stimulating factor (M-CSF) that antagonizes the c-FMS receptor.
Biochem. J., 474, 2017
|
|
6XXO
 
 | |
6XXP
 
 | |
5NX1
 
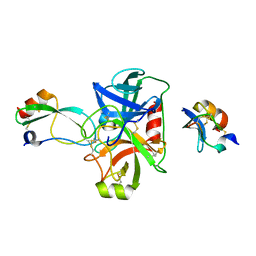 | | Combinatorial Engineering of Proteolytically Resistant APPI Variants that Selectively Inhibit Human Kallikrein 6 for Cancer Therapy | | Descriptor: | Amyloid-beta A4 protein, Kallikrein-6 | | Authors: | Shahar, A, Sananes, A, Radisky, E.S, Papo, N. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | A potent, proteolysis-resistant inhibitor of kallikrein-related peptidase 6 (KLK6) for cancer therapy, developed by combinatorial engineering.
J.Biol.Chem., 293, 2018
|
|
5NX3
 
 | | Combinatorial Engineering of Proteolytically Resistant APPI Variants that Selectively Inhibit Human Kallikrein 6 for Cancer Therapy | | Descriptor: | Amyloid-beta A4 protein, Kallikrein-6 | | Authors: | Shahar, A, Sananes, A, Radisky, E.S, Papo, N. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | A potent, proteolysis-resistant inhibitor of kallikrein-related peptidase 6 (KLK6) for cancer therapy, developed by combinatorial engineering.
J.Biol.Chem., 293, 2018
|
|
7R2W
 
 | | Mutant S-adenosylmethionine synthetase from E.coli complexed with AMPPNP and methionine | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
7R3B
 
 | | S-adenosylmethionine synthetase from Lactobacillus plantarum complexed with AMPPNP, methionine and SAM | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
3W93
 
 | |
3W8V
 
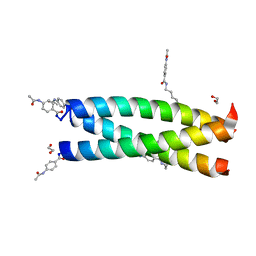 | | Crystal Structure Analysis of the synthetic GCN4 coiled coil peptide | | Descriptor: | 1,2-ETHANEDIOL, GCN4n coiled coil peptide, PARA ACETAMIDO BENZOIC ACID | | Authors: | Shahar, A, Zarivach, R, Ashkenasy, G. | | Deposit date: | 2013-03-21 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-resolution structure that provides insight into coiled-coil thiodepsipeptide dynamic chemistry
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3W92
 
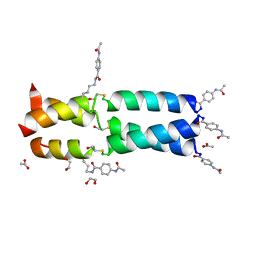 | | Crystal Structure Analysis of the synthetic GCN4 Thioester coiled coil peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PARA ACETAMIDO BENZOIC ACID, ... | | Authors: | Shahar, A, Zarivach, R, Ashkenasy, G. | | Deposit date: | 2013-03-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A high-resolution structure that provides insight into coiled-coil thiodepsipeptide dynamic chemistry
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6GFI
 
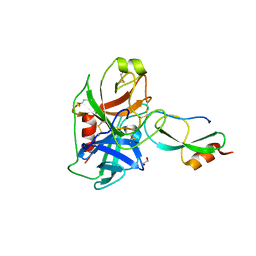 | | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, PRSS3 protein | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N, Naftaly, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.
Nat Commun, 9, 2018
|
|
6RS1
 
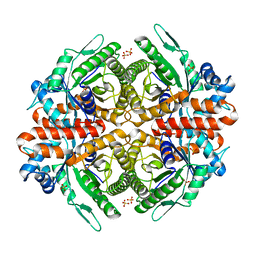 | |
6RNG
 
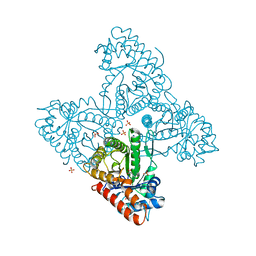 | | Dipeptide Gly-Pro binds to a glycolytic enzyme fructose bisphosphate aldolase | | Descriptor: | Fructose-bisphosphate aldolase 6, cytosolic, GLYCINE, ... | | Authors: | Shahar, A, Zarivach, R, Skirycz, A, Wojciechowska, I. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dipeptide Gly-Pro binds to a glycolytic enzyme fructose bisphosphate aldolase
Not Published
|
|
6RKA
 
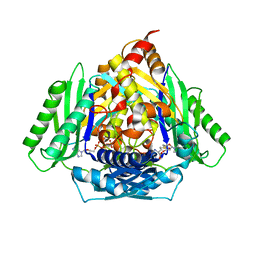 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Methionine adenosyltransferase, PHOSPHATE ION, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RJS
 
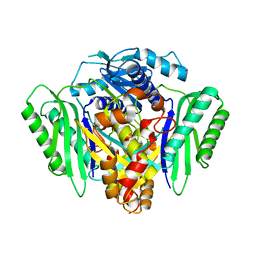 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RK5
 
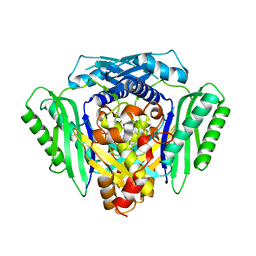 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RK7
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | CHLORIDE ION, Methionine adenosyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RKC
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
7O85
 
 | | Anthrax toxin prepore in complex with the neutralizing Fab cAb29 | | Descriptor: | CALCIUM ION, Fab, Protective antigen PA-63 | | Authors: | Hoelzgen, F, Zalk, R, Alcalay, R, Cohen-Schwartz, S, Garau, G, Shahar, A, Mazor, O, Frank, G.A. | | Deposit date: | 2021-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of the anthrax toxin by antibody-mediated stapling of its membrane-penetrating loop.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7ZVD
 
 | | K89 acetylated glucose-6-phosphate dehydrogenase (G6PD) in a complex with structural NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, F, Muskat, N.H, Nudelman, H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
7ZVE
 
 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
4X71
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Shahar, A, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
