2M70
 
 | |
4U39
 
 | | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis | | Descriptor: | Cell division factor, Cell division protein FtsZ, PHOSPHATE ION | | Authors: | Bisson-Filho, A.W, Discola, K.F, Castellen, P, Blasios, V, Martins, A, Sforca, M.L, Garcia, W, Zeri, A.C, Erickson, H.P, Dessen, A, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis
To be Published
|
|
2POA
 
 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
2PXG
 
 | | NMR Solution Structure of OmlA | | Descriptor: | Outer membrane protein | | Authors: | Vanini, M.M.T, Pertinhez, T.A, Sforca, M.L, Spisni, A, Benedetti, C.E. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. citri reveals a protein fold implicated in protein-protein interaction.
Proteins, 71, 2008
|
|
2L8A
 
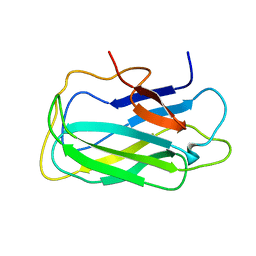 | | Structure of a novel CBM3 lacking the calcium-binding site | | Descriptor: | Endoglucanase | | Authors: | Paiva, J.H, Meza, A.N, Sforca, M.L, Navarro, R.Z, Neves, J.L, Santos, C.R, Murakami, M.T, Zeri, A.C. | | Deposit date: | 2011-01-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | SOLUTION NMR | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
2JP6
 
 | | Structural and functional characterization of the recombinant form of the Kv1.3 channel blocker Tc32 | | Descriptor: | Potassium channel toxin alpha-KTx 18.1 | | Authors: | Stehling, E.G, Sforca, M.L, Zanchin, N.I, Pignatelli, A, Belluzzi, O, Spisni, A, Pertinhez, T.A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the recombinant form of the
Kv1.3 channel blocker Tc32
To be Published
|
|
2JR1
 
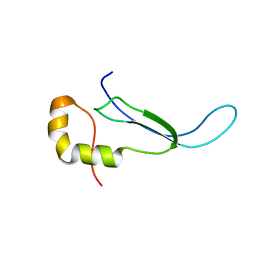 | | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from the phytopathogen Xylella fastidiosa. | | Descriptor: | Virulence regulator | | Authors: | Rosselli, L.K, Sforca, M.L, Souza, A.P, Zeri, A.C. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from the phytopathogen Xylella fastidiosa.
To be Published
|
|
2KDO
 
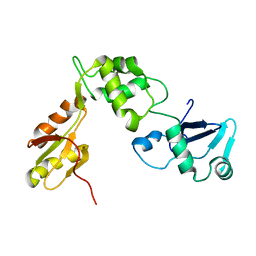 | | Structure of the human Shwachman-Bodian-Diamond syndrome protein, SBDS | | Descriptor: | Ribosome maturation protein SBDS | | Authors: | de Oliveira, J.F, Sforca, M.L, Blumenschein, T, Guimaraes, B.G, Zanchin, N.I.T, Zeri, A.C. | | Deposit date: | 2009-01-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and RNA interaction analysis of the human SBDS protein.
J.Mol.Biol., 396, 2010
|
|
2KQ5
 
 | |
2M4I
 
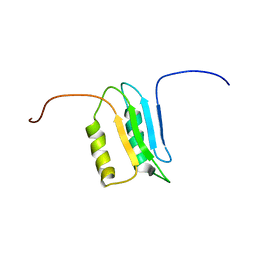 | |
2MRW
 
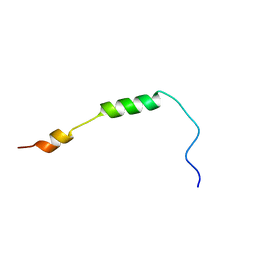 | | Solution Structure of MciZ from Bacillus subtilis | | Descriptor: | Cell division factor | | Authors: | Castellen, P, Sforca, M.L, Zeri, A.C.M, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-16 | | Release date: | 2015-03-25 | | Last modified: | 2015-05-13 | | Method: | SOLUTION NMR | | Cite: | FtsZ filament capping by MciZ, a developmental regulator of bacterial division.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WN5
 
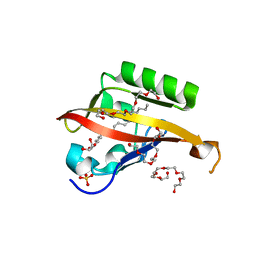 | | Crystal structure of the C-terminal Per-Arnt-Sim (PASb) of human HIF-3alpha9 bound to 18:1-1-monoacylglycerol | | Descriptor: | HEXAETHYLENE GLYCOL, Hypoxia-inducible factor 3-alpha, MONOVACCENIN, ... | | Authors: | Fala, A.M, Oliveira, J.F, Dias, S.M, Ambrosio, A.L. | | Deposit date: | 2014-10-10 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the Hypoxia-inducible factor 3 alpha.
Sci Rep, 5, 2015
|
|
2MMV
 
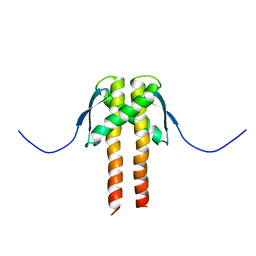 | |
3PZU
 
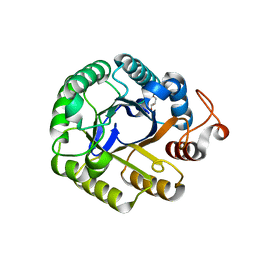 | | P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase, GLYCEROL | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3PZT
 
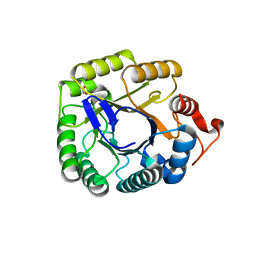 | | Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion | | Descriptor: | Endoglucanase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3PZV
 
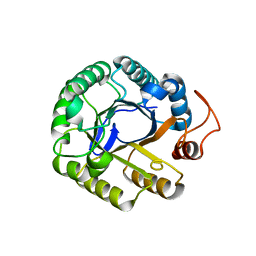 | | C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
4JJM
 
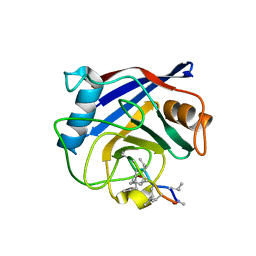 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
6CYR
 
 | |
6CYO
 
 | |
7UXZ
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXX
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
3SUK
 
 | | Crystal structure of cerato-platanin 2 from M. perniciosa (MpCP2) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUJ
 
 | | Crystal structure of cerato-platanin 1 from M. perniciosa (MpCP1) | | Descriptor: | ACETATE ION, CHLORIDE ION, Cerato-platanin 1, ... | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUM
 
 | | Crystal structure of cerato-platanin 5 from M. perniciosa (MpCP5) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
