5K99
 
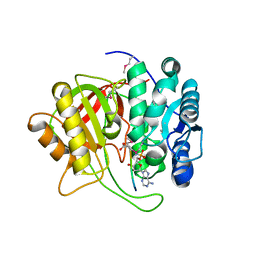 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
1HQM
 
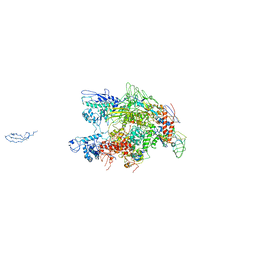 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2MW3
 
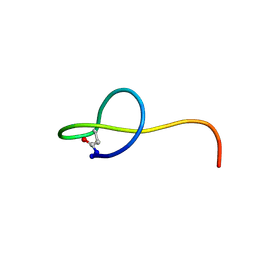 | | Solution NMR structure of the lasso peptide streptomonomicin | | Descriptor: | Lasso peptide | | Authors: | Tietz, J.I, Zhu, L, Mitchell, D.A, Metelev, M, Melby, J.O, Blair, P.M, Livnat, I, Severinov, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-01-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete.
Chem.Biol., 22, 2015
|
|
4ZJN
 
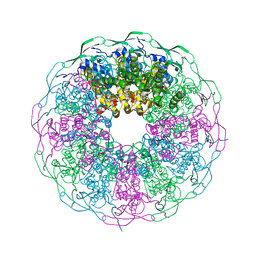 | | Crystal structure of the bacteriophage G20C portal protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Portal protein | | Authors: | Williams, L.S, Turkenburg, J.P, Levdikov, V.M, Minakhin, L, Severinov, K, Antson, A.A. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the bacteriophage G20C portal protein
To be published
|
|
4E94
 
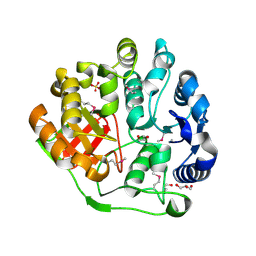 | | Crystal structure of MccF-like protein from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, MccC family protein, SULFATE ION | | Authors: | Nocek, B, Tikhonov, A, Kwon, K, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of MccF-like protein from Streptococcus pneumoniae
TO BE PUBLISHED
|
|
6U48
 
 | | E. coli 50S with phazolicin (PHZ) bound in exit tunnel | | Descriptor: | 23S rRNA, 50S ribosomal protein bL17, 50S ribosomal protein bL19, ... | | Authors: | Watson, Z.L, Cate, J.H.D, Polikanov, Y.S, Severinov, K. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition.
Nat Commun, 10, 2019
|
|
1YNN
 
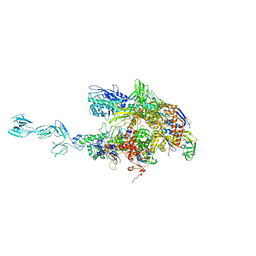 | | Taq RNA polymerase-rifampicin complex | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Campbell, E.A, Pavlova, O, Zenkin, N, Leon, F, Irschik, H, Jansen, R, Severinov, K, Darst, S.A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Embo J., 24, 2005
|
|
4MI1
 
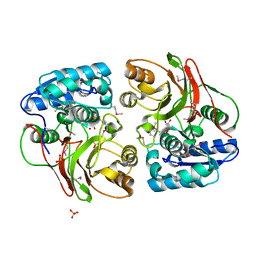 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Microcin immunity protein MccF, SULFATE ION | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
5W4K
 
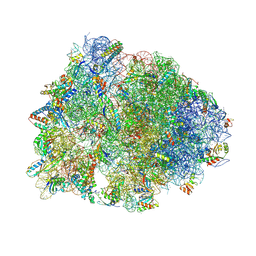 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | Descriptor: | Klebsidin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
5UI6
 
 | | Solution NMR Structure of Lasso Peptide Acinetodin | | Descriptor: | Acinetodin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
1YNJ
 
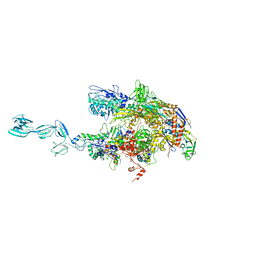 | | Taq RNA polymerase-Sorangicin complex | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Campbell, E.A, Pavlova, O, Zenkin, N, Leon, F, Irschik, H, Jansen, R, Severinov, K, Darst, S.A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Embo J., 24, 2005
|
|
6GRG
 
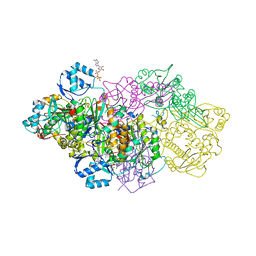 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17, ADP and phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GRI
 
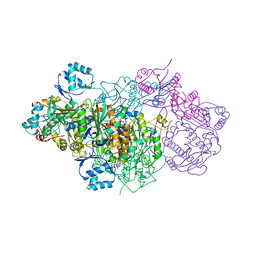 | | E. coli Microcin synthetase McbBCD complex | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Microcin B17-processing protein McbB, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GRH
 
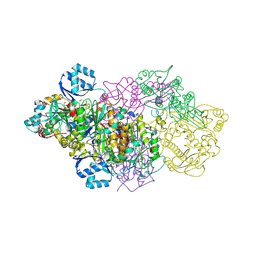 | | E. coli Microcin synthetase McbBCD complex with truncated pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6GOS
 
 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
5F5X
 
 | |
5F9M
 
 | | Crystal structure of native B3275, member of MccF family of enzymes | | Descriptor: | MccC family protein | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of native B3275, member of MccF family of enzymes.
To Be Published
|
|
5FCD
 
 | | Crystal structure of MccD protein | | Descriptor: | CHLORIDE ION, MccD, UNK-UNK-UNK-MSE-UNK, ... | | Authors: | Nocek, B, Jedrzejczak, R, Anderson, W.F, Severinov, K, Dubiley, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MccD protein.
To Be Published
|
|
5FD8
 
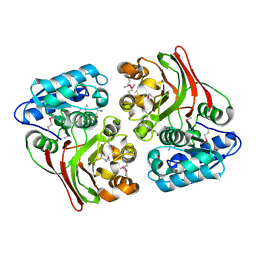 | |
2MC5
 
 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by -factor displacement | | Descriptor: | 45L | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2014-04-30 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
2MC6
 
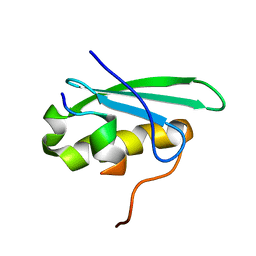 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement | | Descriptor: | DNA-directed RNA polymerase subunit beta', RNA polymerase inhibitor p7 | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-15 | | Release date: | 2014-04-02 | | Last modified: | 2014-05-07 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
4INJ
 
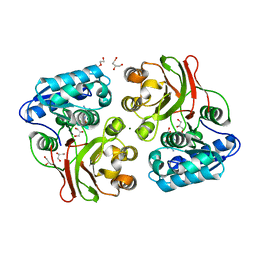 | | Crystal structure of the S111A mutant of member of MccF clade from Listeria monocytogenes EGD-e with product | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1638 protein, ... | | Authors: | Nocek, B, Tikhonov, A, Severinov, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the S111A mutant of member of MccF clade from Listeria monocytogenes EGD-e with product
To be Published
|
|
4JVO
 
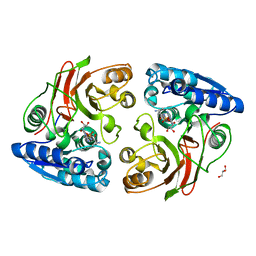 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Microcin immunity protein MccF | | Authors: | Nocek, B, Tikhonov, A, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
2A6H
 
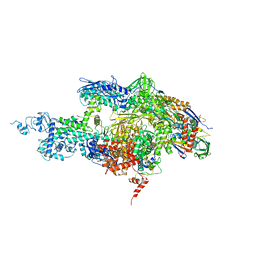 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Temiakov, D, Zenkin, N, Vassylyeva, M.N, Perederina, A, Tahirov, T.H, Savkina, M, Zorov, S, Nikiforov, V, Igarashi, N, Matsugaki, N, Wakatsuki, S, Severinov, K, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of transcription inhibition by antibiotic streptolydigin.
Mol.Cell, 19, 2005
|
|
