1A44
 
 | | PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN FROM CALF BRAIN | | Descriptor: | ACETATE ION, PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN | | Authors: | Serre, L, Vallee, B, Zelwer, C, Schoentgen, F. | | Deposit date: | 1998-02-10 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins.
Structure, 6, 1998
|
|
1B7A
 
 | | STRUCTURE OF THE PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN FROM BOVINE BRAIN | | Descriptor: | PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Serre, L, Vallee, B, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 1999-01-21 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins.
Structure, 6, 1998
|
|
1MLA
 
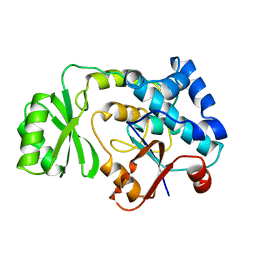 | | THE ESCHERICHIA COLI MALONYL-COA:ACYL CARRIER PROTEIN TRANSACYLASE AT 1.5-ANGSTROMS RESOLUTION. CRYSTAL STRUCTURE OF A FATTY ACID SYNTHASE COMPONENT | | Descriptor: | MALONYL-COENZYME A ACYL CARRIER PROTEIN TRANSACYLASE | | Authors: | Serre, L, Verbree, E.C, Dauter, Z, Stuitje, A.R, Derewenda, Z.S. | | Deposit date: | 1995-01-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5-A resolution. Crystal structure of a fatty acid synthase component.
J.Biol.Chem., 270, 1995
|
|
8C5C
 
 | |
1QUE
 
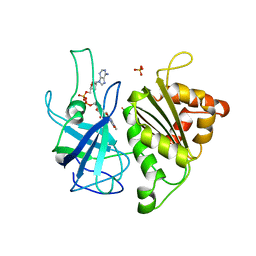 | | X-RAY STRUCTURE OF THE FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 1.8 ANGSTROMS | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-07-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
1QUF
 
 | | X-RAY STRUCTURE OF A COMPLEX NADP+-FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 2.25 ANGSTROMS | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-09-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
1KFV
 
 | | Crystal Structure of Lactococcus lactis Formamido-pyrimidine DNA Glycosylase (alias Fpg or MutM) Non Covalently Bound to an AP Site Containing DNA. | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*C)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*G)-3', Formamido-pyrimidine DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA.
EMBO J., 21, 2002
|
|
1CJX
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS FLUORESCENS HPPD | | Descriptor: | 4-HYDROXYPHENYLPYRUVATE DIOXYGENASE, ACETATE ION, ETHYL MERCURY ION, ... | | Authors: | Serre, L, Sailland, A, Sy, D, Boudec, P, Rolland, A, Pebay-Peroulla, E, Cohen-Addad, C. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway.
Structure Fold.Des., 7, 1999
|
|
1F4L
 
 | | CRYSTAL STRUCTURE OF THE E.COLI METHIONYL-TRNA SYNTHETASE COMPLEXED WITH METHIONINE | | Descriptor: | METHIONINE, METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Serre, L, Verdon, G, Chonowski, T, Hervouet, N, Zelwer, C. | | Deposit date: | 2000-06-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding.
J.Mol.Biol., 306, 2001
|
|
1FJJ
 
 | | CRYSTAL STRUCTURE OF E.COLI YBHB PROTEIN, A NEW MEMBER OF THE MAMMALIAN PEBP FAMILY | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HYPOTHETICAL 17.1 KDA PROTEIN IN MODC-BIOA INTERGENIC REGION | | Authors: | Serre, L, Pereira de Jesus, K, Benedetti, H, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 2000-08-08 | | Release date: | 2001-07-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein.
J.Mol.Biol., 310, 2001
|
|
1FUX
 
 | | CRYSTAL STRUCTURE OF E.COLI YBCL, A NEW MEMBER OF THE MAMMALIAN PEBP FAMILY | | Descriptor: | HYPOTHETICAL 19.5 KDA PROTEIN IN EMRE-RUS INTERGENIC REGION | | Authors: | Serre, L, Pereira de Jesus, K, Benedetti, H, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 2000-09-18 | | Release date: | 2001-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein.
J.Mol.Biol., 310, 2001
|
|
1OSD
 
 | | crystal structure of Oxidized MerP from Ralstonia metallidurans CH34 | | Descriptor: | hypothetical protein MerP | | Authors: | Serre, L, Rossy, E, Pebay-Peyroula, E, Cohen-Addad, C, Coves, J. | | Deposit date: | 2003-03-19 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxidized Form of the Periplasmic Mercury-binding Protein MerP from Ralstonia metallidurans CH34
J.MOL.BIOL., 339, 2004
|
|
1NNJ
 
 | | Crystal structure Complex between the Lactococcus lactis Fpg and an abasic site containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1PJJ
 
 | | Complex between the Lactococcus lactis Fpg and an abasic site containing DNA. | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1U3A
 
 | | mutant DsbA | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Thiol: disulfide interchange protein dsbA | | Authors: | Serre, L. | | Deposit date: | 2004-07-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant
J.Mol.Biol., 347, 2005
|
|
3HZ8
 
 | | Crystal structure of the oxidized T176V DsbA1 mutant | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Iwema, T, Carpentier, P, Jullian-Binard, C, Serre, L. | | Deposit date: | 2009-06-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
1WAB
 
 | | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | ACETATE ION, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Swenson, L, Derewenda, U, Serre, L, Wei, Y, Dauter, Z, Hattori, M, Aoki, J, Arai, H, Adachi, T, Inoue, K, Derewenda, Z.S. | | Deposit date: | 1996-10-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer.
Nature, 385, 1997
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1PM5
 
 | | Crystal structure of wild type Lactococcus lactis Fpg complexed to a tetrahydrofuran containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira de Jesus-Tran, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
2B3S
 
 | |
2B6M
 
 | |
1TI1
 
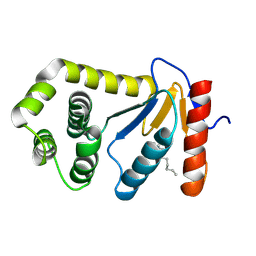 | | crystal structure of a mutant DsbA | | Descriptor: | DODECANE, Thiol:disulfide interchange protein dsbA | | Authors: | Kone, A, Serre, L. | | Deposit date: | 2004-06-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant.
J.Mol.Biol., 347, 2005
|
|
3DVW
 
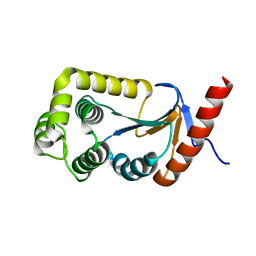 | |
3DVX
 
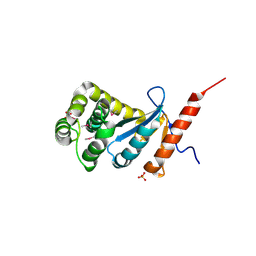 | | Crystal structure of reduced DsbA3 from Neisseria meningitidis | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Serre, L. | | Deposit date: | 2008-07-21 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
1PJI
 
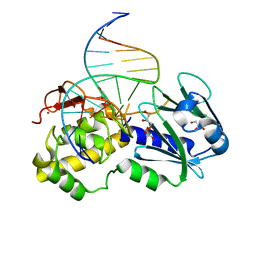 | | Crystal structure of wild type Lactococcus lactis FPG complexed to a 1,3 propanediol containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
