2GDI
 
 | |
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKQ
 
 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YLS
 
 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1Y27
 
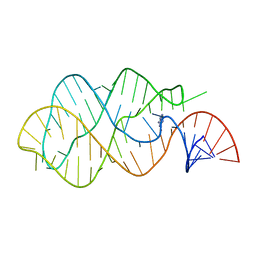 | | G-riboswitch-guanine complex | | Descriptor: | Bacillus subtilis xpt, GUANINE | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
1Y26
 
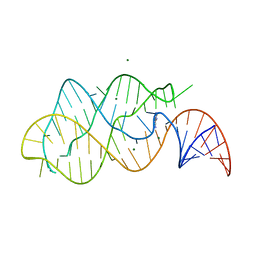 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
4GXY
 
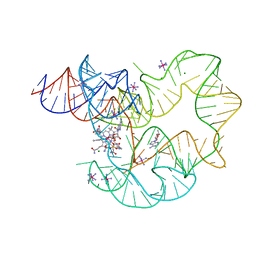 | | RNA structure | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Serganov, A, Peselis, A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1AB3
 
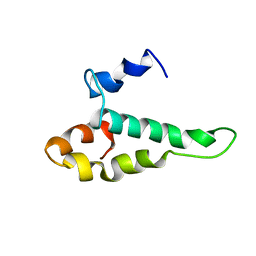 | | RIBOSOMAL PROTEIN S15 FROM THERMUS THERMOPHILUS, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL RNA BINDING PROTEIN S15 | | Authors: | Berglund, H, Rak, A, Serganov, A, Garber, M, Hard, T. | | Deposit date: | 1997-02-03 | | Release date: | 1997-04-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
8FVR
 
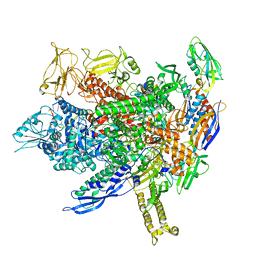 | | CryoEM structure of E.coli transcription elongation complex | | Descriptor: | DNA (53-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Duan, W, Serganov, A. | | Deposit date: | 2023-01-19 | | Release date: | 2023-04-05 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Control of transcription elongation and DNA repair by alarmone ppGpp.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FVW
 
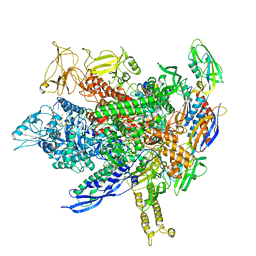 | |
2LA5
 
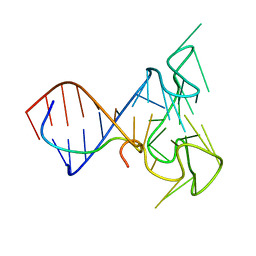 | | RNA Duplex-Quadruplex Junction Complex with FMRP RGG peptide | | Descriptor: | Fragile X mental retardation 1 protein, RNA (36-MER) | | Authors: | Phan, A, Kuryavyi, V, Darnell, J, Serganov, A, Majumdar, A, Ilin, S, Darnell, R, Patel, D. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-20 | | Method: | SOLUTION NMR | | Cite: | Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1F7Y
 
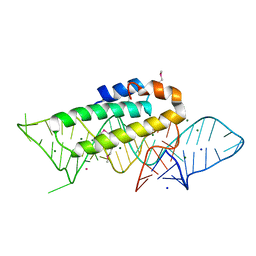 | | THE CRYSTAL STRUCTURE OF TWO UUCG LOOPS HIGHLIGHTS THE ROLE PLAYED BY 2'-HYDROXYL GROUPS IN ITS UNUSUAL STABILITY | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Nikouline, A, Serganov, A, Tishchenko, S, Nevskaya, N, Garber, M, Ehresmann, B, Ehresmann, C, Nikonov, S, Dumas, P. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of UUCG tetraloop.
J.Mol.Biol., 304, 2000
|
|
2VAZ
 
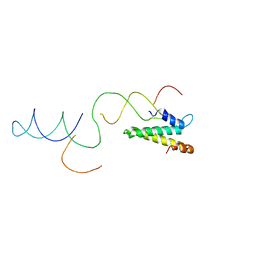 | | Model of the S15-mRNA complex fitted into the cryo-EM map of the 70S entrapment complex. | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, RPSO MRNA OPERATOR | | Authors: | Marzi, S, Myasnikov, A.G, Serganov, A, Ehresmann, C, Romby, P, Yusupov, M, Klaholz, B.P. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-02 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structured Mrnas Regulate Translation Initiation by Binding to the Platform of the Ribosome.
Cell(Cambridge,Mass.), 130, 2007
|
|
1DK1
 
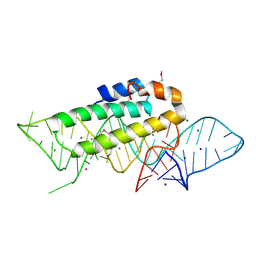 | | DETAILED VIEW OF A KEY ELEMENT OF THE RIBOSOME ASSEMBLY: CRYSTAL STRUCTURE OF THE S15-RRNA COMPLEX | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Nikulin, A, Serganov, A, Ennifar, E, Tischenko, S, Nevskaya, N. | | Deposit date: | 1999-12-06 | | Release date: | 2000-04-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S15-rRNA complex.
Nat.Struct.Biol., 7, 2000
|
|
7TZT
 
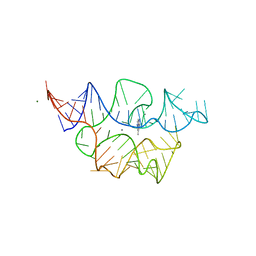 | | Crystal structure of the E. coli thiM riboswitch in complex with N1,N1-dimethyl-N2-(quinoxalin-6-ylmethyl)ethane-1,2-diamine (linked compound 37) | | Descriptor: | 6-methylquinoxaline, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | SHAPE-enabled fragment-based ligand discovery for RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TZR
 
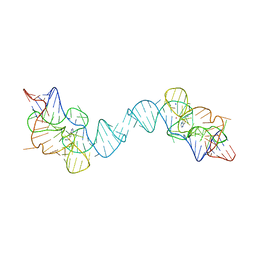 | |
7TZU
 
 | |
7TZS
 
 | |
7TD7
 
 | |
7TDB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, manganese ions | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TDC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, calcium ions | | Descriptor: | CALCIUM ION, MAGNESIUM ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TDA
 
 | |
7MD9
 
 | |
7MD6
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL1 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MDB
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CACODYLATE ION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
