1F6Y
 
 | |
1MJG
 
 | | CRYSTAL STRUCTURE OF BIFUNCTIONAL CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE(CODH/ACS) FROM MOORELLA THERMOACETICA (F. CLOSTRIDIUM THERMOACETICUM) | | Descriptor: | ACETATE ION, CARBON MONOXIDE DEHYDROGENASE BETA SUBUNIT, COPPER (I) ION, ... | | Authors: | Doukov, T.I, Iverson, T.M, Seravalli, J, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2002-08-27 | | Release date: | 2003-01-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA
synthase
Science, 298, 2002
|
|
5UCU
 
 | | STRUCTURAL AND MECHANISTIC INSIGHTS INTO HEMOGLOBIN-CATALYZED HYDROGEN SULFIDE OXIDATION AND THE FATE OF POLYSULFIDE PRODUCTS | | Descriptor: | HYDROSULFURIC ACID, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Vitvitsky, V, Yadav, P.K, An, S, Seravalli, J, Cho, U.-S, Banerjee, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural and Mechanistic Insights into Hemoglobin-catalyzed Hydrogen Sulfide Oxidation and the Fate of Polysulfide Products.
J. Biol. Chem., 292, 2017
|
|
6UND
 
 | |
6UNF
 
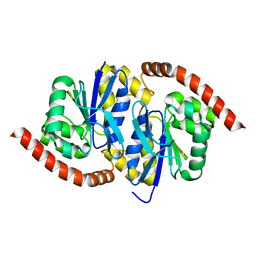 | |
8DKG
 
 | | Structure of PYCR1 Thr171Met variant complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Isoform 3 of Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Meeks, K.R, Tanner, J.J. | | Deposit date: | 2022-07-05 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional Impact of a Cancer-Related Variant in Human Delta 1 -Pyrroline-5-Carboxylate Reductase 1.
Acs Omega, 8, 2023
|
|
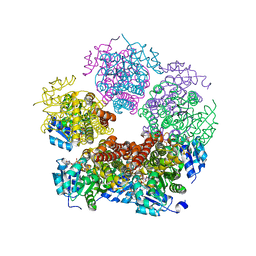
2Z8Y
 
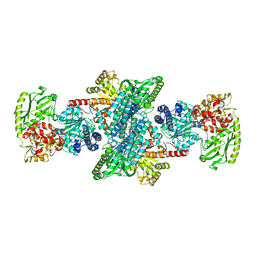 | | Xenon-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase(CODH/ACS) from Moorella thermoacetica | | Descriptor: | COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl CoA synthase subunit alpha, Carbon monoxide dehydrogenase/acetyl CoA synthase subunit beta, ... | | Authors: | Doukov, T.I, Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Xenon in and at the End of the Tunnel of Bifunctional Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Biochemistry, 47, 2008
|
|
6NI4
 
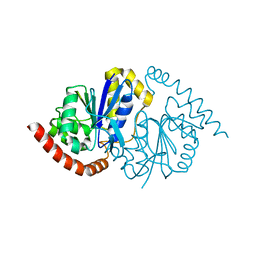 | |
6NI7
 
 | |
6NPQ
 
 | |
6NIA
 
 | |
6NI9
 
 | |
6NI5
 
 | |
6NI6
 
 | |
3I01
 
 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | Descriptor: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
3I04
 
 | | Cyanide-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, cyanide-bound C-cluster | | Descriptor: | ACETATE ION, COPPER (I) ION, CYANIDE ION, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
5VPS
 
 | | Crystal structure of an SDR from Burkholderia ambifaria in complex with NADPH with a TCEP adduct | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase/reductase SDR, [(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]piperidin-4-yl]-tris(3-hydroxy-3-oxopropyl)phosphanium | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Cautionary Tale of Using Tris(alkyl)phosphine Reducing Agents with NAD+-Dependent Enzymes.
Biochemistry, 2020
|
|
4DJD
 
 | | Crystal structure of folate-free corrinoid iron-sulfur protein (CFeSP) in complex with its methyltransferase (MeTr) | | Descriptor: | 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION, COBALAMIN, ... | | Authors: | Kung, Y, Doukov, T.I, Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Visualizing molecular juggling within a B12-dependent methyltransferase complex.
Nature, 484, 2012
|
|
4DJF
 
 | | Crystal structure of folate-bound corrinoid iron-sulfur protein (CFeSP) in complex with its methyltransferase (MeTr), co-crystallized with folate and Ti(III) citrate reductant | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION, ... | | Authors: | Kung, Y, Drennan, C.L. | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Visualizing molecular juggling within a B12-dependent methyltransferase complex.
Nature, 484, 2012
|
|
4DJE
 
 | | Crystal structure of folate-bound corrinoid iron-sulfur protein (CFeSP) in complex with its methyltransferase (MeTr), co-crystallized with folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION, ... | | Authors: | Kung, Y, Drennan, C.L. | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.504 Å) | | Cite: | Visualizing molecular juggling within a B12-dependent methyltransferase complex.
Nature, 484, 2012
|
|
6NI8
 
 | |
2E7F
 
 | | 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.2 Angsrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
2OGY
 
 | | Asn199Ala Mutant of the 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.3 Angstrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
