1A7X
 
 | | FKBP12-FK1012 COMPLEX | | Descriptor: | BENZYL-CARBAMIC ACID [8-DEETHYL-ASCOMYCIN-8-YL]ETHYL ESTER, FKBP12 | | Authors: | Schultz, L.W, Clardy, J. | | Deposit date: | 1998-03-18 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical inducers of dimerization: the atomic structure of FKBP12-FK1012A-FKBP12.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
4NNR
 
 | | FKBP13-FK506 Complex | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP2 | | Authors: | Schultz, L.W, Martin, P.K, Liang, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 2013-11-18 | | Release date: | 2014-02-05 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Atomic structure of the Immunophilin FKBP13-FK506 Complex: Insights into the Composite Binding Surface for Calcineurin
J.Am.Chem.Soc., 116, 1994
|
|
1VPR
 
 | |
3RSP
 
 | | STRUCTURE OF THE P93G VARIANT OF RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Schultz, L.W, Hargraves, S.R, Klink, T.A, Raines, R.T. | | Deposit date: | 1997-10-20 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of the P93G variant of ribonuclease A.
Protein Sci., 7, 1998
|
|
3RSD
 
 | |
4RSD
 
 | | STRUCTURE OF THE D121A VARIANT OF RIBONUCLEASE A | | Descriptor: | ACETATE ION, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Schultz, L.W, Quirk, D.J, Raines, R.T. | | Deposit date: | 1998-02-05 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | His...Asp catalytic dyad of ribonuclease A: structure and function of the wild-type, D121N, and D121A enzymes.
Biochemistry, 37, 1998
|
|
1TXX
 
 | |
3RSK
 
 | |
4RSK
 
 | |
4E4V
 
 | | The crystal structure of the dimeric human importin alpha 1 at 2.5 angstrom resolution. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Importin subunit alpha-2 | | Authors: | Hang, P.C, Miknis, Z.M, Franke, W.A, Umland, T.C, Schultz, L.W. | | Deposit date: | 2012-03-13 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5283 Å) | | Cite: | The crystal structure of the dimeric human importin alpha 1 at 2.5 angstrom resolution.
To be Published
|
|
1C8W
 
 | | THR45GLY VARIANT OF RIBONUCLEASE A | | Descriptor: | ACETATE ION, CHLORIDE ION, PROTEIN (Ribonuclease A) | | Authors: | Kelemen, B.R, Sweeney, R.T, Schultz, L.W, Raines, R.T. | | Deposit date: | 1999-07-30 | | Release date: | 2002-05-01 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Excavating an active site: the nucleobase specificity of ribonuclease A.
Biochemistry, 39, 2000
|
|
1C9X
 
 | | H119A VARIANT OF RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Park, C, Schultz, L.W, Raines, R.T. | | Deposit date: | 1999-08-03 | | Release date: | 2001-06-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the active site histidine residues of ribonuclease A to nucleic acid binding.
Biochemistry, 40, 2001
|
|
1C9V
 
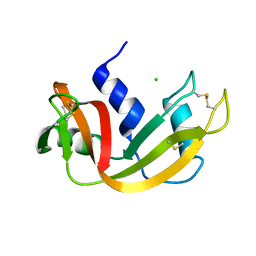 | | H12A VARIANT OF RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Park, C, Schultz, L.W, Raines, R.T. | | Deposit date: | 1999-08-03 | | Release date: | 2001-06-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the active site histidine residues of ribonuclease A to nucleic acid binding.
Biochemistry, 40, 2001
|
|
4Y0A
 
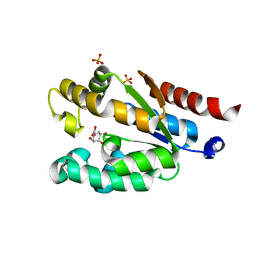 | | Shikimate kinase from Acinetobacter baumannii in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, Shikimate kinase | | Authors: | Sutton, K.A, Breen, J, MacDonald, U, Beanan, J.M, Olson, R, Russo, T.A, Schultz, L.W, Umland, T.C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structure of shikimate kinase, an in vivo essential metabolic enzyme in the nosocomial pathogen Acinetobacter baumannii, in complex with shikimate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3EE7
 
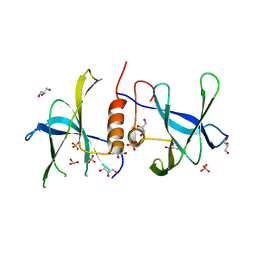 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
1FKI
 
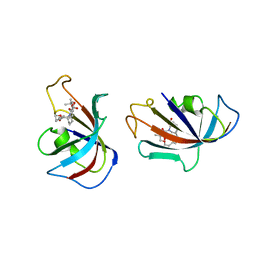 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | (21S)-1AZA-4,4-DIMETHYL-6,19-DIOXA-2,3,7,20-TETRAOXOBICYCLO[19.4.0] PENTACOSANE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1FKG
 
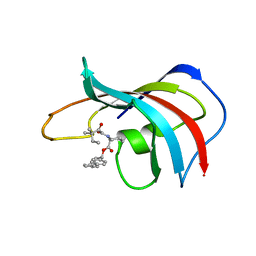 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1,3-DIPHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1FKH
 
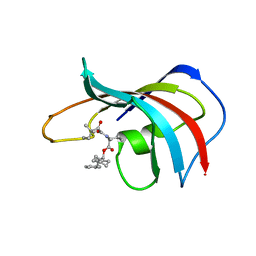 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
5BS5
 
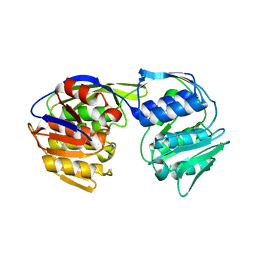 | | EPSP synthase from Acinetobacter baumannii | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Sutton, K.A, Schultz, L.W, Russo, T.A, Breen, J, Umland, T.C. | | Deposit date: | 2015-06-01 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | EPSP synthase from Acinetobacter baumannii
Acta Crystallogr.,Sect.F, 2016
|
|
5BUF
 
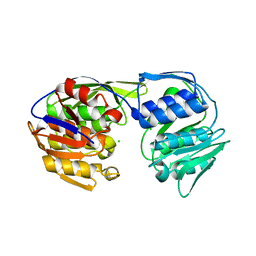 | | 2.37 Angstrom Structure of EPSP Synthase from acinetobacter baumannii | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION | | Authors: | Sutton, K.A, Schultz, L.W, Breen, J, Graham, J, Umland, T.C. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase from the ESKAPE pathogen Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1F40
 
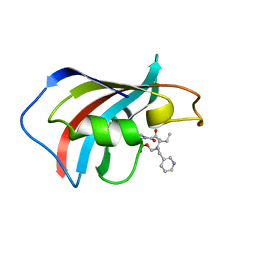 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1N3Z
 
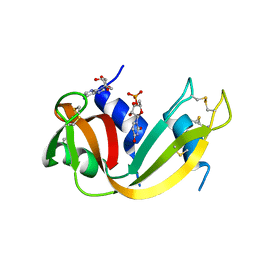 | | Crystal structure of the [S-carboxyamidomethyl-Cys31, S-carboxyamidomethyl-Cys32] monomeric derivative of the bovine seminal ribonuclease in the liganded state | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, ADENOSINE, Ribonuclease, ... | | Authors: | Sica, F, Di Fiore, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-10-30 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative
Proteins, 52, 2003
|
|
