2CF4
 
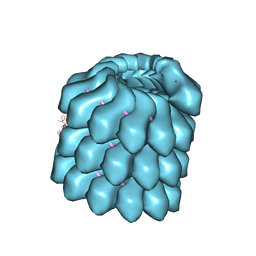
 | | Pyrococcus horikoshii TET1 peptidase can assemble into a tetrahedron or a large octahedral shell | | Descriptor: | COBALT (II) ION, PROTEIN PH0519 | | Authors: | Vellieux, F.M.D, Schoehn, G, Dura, M.A, Roussel, A, Franzetti, B. | | Deposit date: | 2006-02-15 | | Release date: | 2006-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | An Archaeal Peptidase Assembles Into Two Different Quaternary Structures: A Tetrahedron and a Giant Octahedron.
J.Biol.Chem., 281, 2006
|
|
7Q1Z
 
 | | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Sulbaran, G, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.
Cell Rep Med, 3, 2022
|
|
5G2E
 
 | | Structure of the Nap1 H2A H2B complex | | Descriptor: | HISTONE H2A TYPE 1, HISTONE H2B 1.1, NUCLEOSOME ASSEMBLY PROTEIN | | Authors: | AguilarGurrieri, C, Larabi, A, Vinayachandran, V, Patel, N.A, Yen, K, Reja, R, Ebong, I.O, Schoehn, G, Robinson, C.V, Pugh, B.F, Panne, D. | | Deposit date: | 2016-04-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structural Evidence for Nap1-Dependent H2A-H2B Deposition and Nucleosome Assembly.
Embo J., 35, 2016
|
|
3KIP
 
 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5NGJ
 
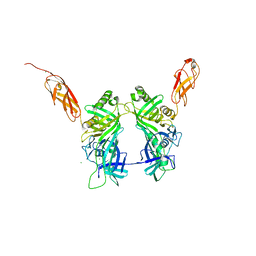 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
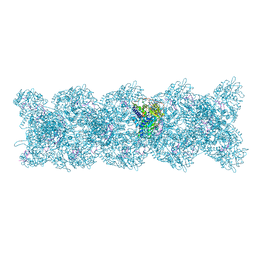
4UFT
 
 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
1H7Z
 
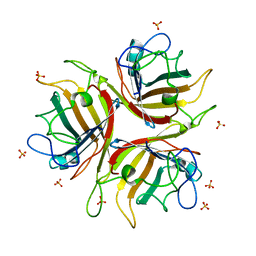 | | Adenovirus Ad3 fibre head | | Descriptor: | ADENOVIRUS FIBRE PROTEIN, SULFATE ION | | Authors: | Durmort, C, Stehlin, C, Schoehn, G, Mitraki, A, Drouet, E, Cusack, S, Burmeister, W.P. | | Deposit date: | 2001-01-21 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Fiber Head of Ad3, a Non-Car-Binding Serotype of Adenovirus
Virology, 285, 2001
|
|
8PZP
 
 | | Model for influenza A virus helical ribonucleoprotein-like structure | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
8PZQ
 
 | | Model for focused reconstruction of influenza A RNP-like particle | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
8CK0
 
 | |
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CPC
 
 | |
4V4U
 
 | | The quasi-atomic model of Human Adenovirus type 5 capsid | | Descriptor: | HEXON PROTEIN, N-TERMINAL PEPTIDE OF FIBER PROTEIN, PENTON PROTEIN | | Authors: | Fabry, C.M.S, Rosa-Calatrava, M, Conway, J.F, Zubieta, C, Cusack, S, Ruigrok, R.W.H, Schoehn, G. | | Deposit date: | 2005-03-03 | | Release date: | 2014-07-09 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | A Quasi-Atomic Model of Human Adenovirus Type 5 Capsid.
Embo J., 24, 2005
|
|
1O7D
 
 | | The structure of the bovine lysosomal a-mannosidase suggests a novel mechanism for low pH activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Heikinheimo, P, Helland, R, Leiros, H.S, Leiros, I, Karlsen, S, Evjen, G, Ravelli, R, Schoehn, G, Ruigrok, R, Tollersrud, O.-K, Mcsweeney, S, Hough, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of Bovine Lysosomal Alpha-Mannosidase Suggests a Novel Mechanism for Low-Ph Activation
J.Mol.Biol., 327, 2003
|
|
2C9F
 
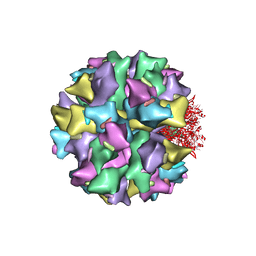 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON DODECAHEDRON | | Descriptor: | FIBER, PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-02 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
2C9G
 
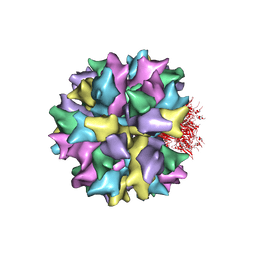 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON BASE DODECAHEDRON | | Descriptor: | PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-04 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
6F3K
 
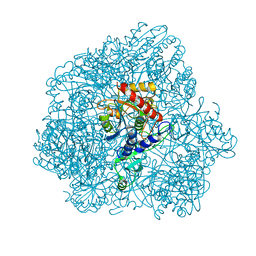 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
7PLM
 
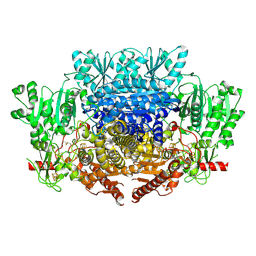 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
1O6E
 
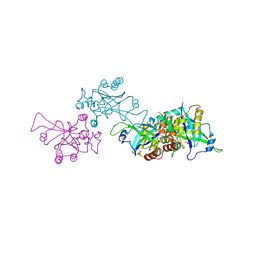 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
7QG9
 
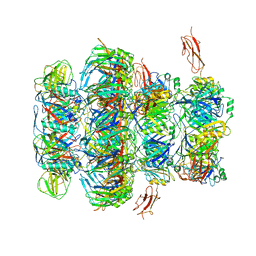 | | Tail tip of siphophage T5 : common core proteins | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7OZ4
 
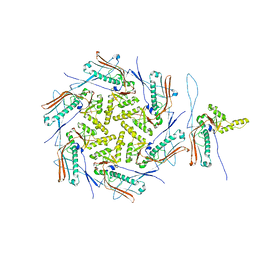 | | Mature capsid of bacteriophage phiRSA1 | | Descriptor: | p2 family phage major capsid protein | | Authors: | Effantin, G, Fujiwara, A, Kawsaki, T, Yamada, T, Schoehn, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High Resolution Structure of the Mature Capsid of Ralstonia solanacearum Bacteriophage phi RSA1 by Cryo-Electron Microscopy.
Int J Mol Sci, 22, 2021
|
|
6GGS
 
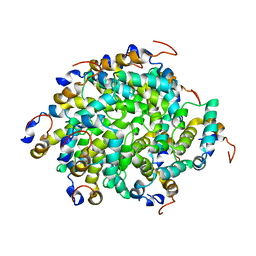 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
1T3E
 
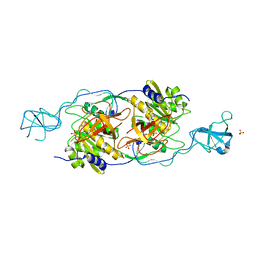 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
5LDF
 
 | |
