6QAY
 
 | |
1EGX
 
 | | SOLUTION STRUCTURE OF THE ENA-VASP HOMOLOGY 1 (EVH1) DOMAIN OF HUMAN VASODILATOR-STIMULATED PHOSPHOPROTEIN (VASP) | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Ball, L, Kuhne, R, Hoffmann, B, Hafner, A, Schmieder, P, Volkmer-Engert, R, Hof, M, Wahl, M, Schneider-Mergener, J, Walter, U, Oschkinat, H, Jarchau, T. | | Deposit date: | 2000-02-17 | | Release date: | 2000-09-20 | | Last modified: | 2012-07-25 | | Method: | SOLUTION NMR | | Cite: | Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity.
EMBO J., 19, 2000
|
|
1XZ9
 
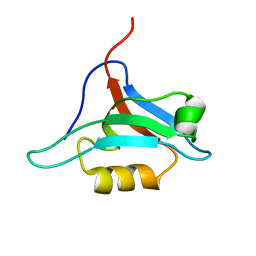 | | Structure of AF-6 PDZ domain | | Descriptor: | Afadin | | Authors: | Joshi, M, Boisguerin, P, Leitner, D, Volkmer-Engert, R, Moelling, K, Schade, M, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2004-11-12 | | Release date: | 2005-11-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1SGG
 
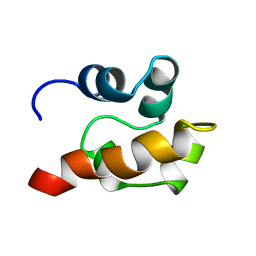 | | THE SOLUTION STRUCTURE OF SAM DOMAIN FROM THE RECEPTOR TYROSINE KINASE EPHB2, NMR, 10 STRUCTURES | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Smalla, M, Schmieder, P, Kelly, M, Ter Laak, A, Krause, G, Ball, L, Wahl, M, Bork, P, Oschkinat, H. | | Deposit date: | 1999-01-08 | | Release date: | 1999-10-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites.
Protein Sci., 8, 1999
|
|
5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N, ethane-1,2-diol | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OF2
 
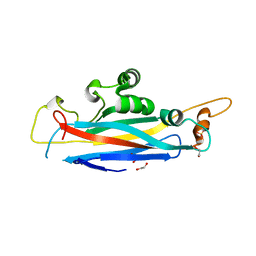 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1SKL
 
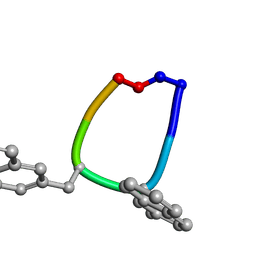 | | Structure of the antimicrobial hexapeptide cyc-(RRNalNalRF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide RR(NAL)(NAL)RF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
5IB3
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, COPPER (II) ION, GLYCEROL, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB4
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Nickel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB1
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR measured at 295 K | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB5
 
 | | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB2
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
2I9S
 
 | | The solution structure of the core of mesoderm development (MESD). | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Andersen, O, Diehl, A, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2JRZ
 
 | | Solution structure of the Bright/ARID domain from the human JARID1C protein. | | Descriptor: | Histone demethylase JARID1C | | Authors: | Koehler, C, Bishop, S, Dowler, E.F, Diehl, A, Schmieder, P, Leidert, M, Sundstrom, M, Arrowsmith, C.H, Wiegelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein.
Biomol.Nmr Assign., 2, 2008
|
|
2MBK
 
 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | Descriptor: | Crossveinless 2 | | Authors: | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | Deposit date: | 2013-08-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
2MWG
 
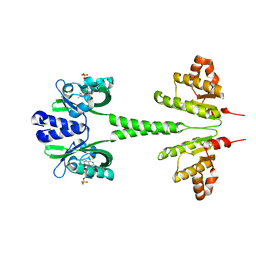 | |
1OYI
 
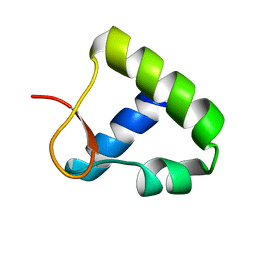 | | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L | | Descriptor: | double-stranded RNA-binding protein | | Authors: | Kahmann, J.D, Wecking, D.A, Putter, V, Lowenhaupt, K, Kim, Y.-G, Schmieder, P, Oschkinat, H, Rich, A, Schade, M. | | Deposit date: | 2003-04-04 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2OTQ
 
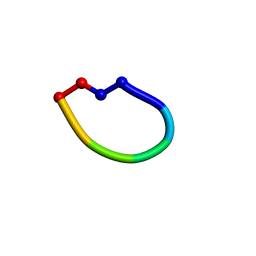 | | Structure of the antimicrobial peptide cyclo(RRWFWR) bound to DPC micelles | | Descriptor: | cRW3 cationic antimicrobial peptide | | Authors: | Appelt, C, Wesselowski, A, Soderhall, J.A, Dathe, M, Schmieder, P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structures of cyclic, antimicrobial peptides in a membrane-mimicking environment define requirements for activity.
J.Pept.Sci., 14, 2007
|
|
2OX2
 
 | | Structure of the cantionic, antimicrobial hexapeptide cyclo(RRWWFR) bound to DPC-micelles | | Descriptor: | cRW2 peptide | | Authors: | Appelt, C, Wessolowski, A, Soderhall, J.A, Dathe, M, Schmieder, P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structures of cyclic, antimicrobial peptides in a membrane-mimicking environment define requirements for activity.
J.Pept.Sci., 14, 2007
|
|
2PDZ
 
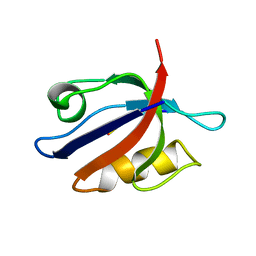 | | SOLUTION STRUCTURE OF THE SYNTROPHIN PDZ DOMAIN IN COMPLEX WITH THE PEPTIDE GVKESLV, NMR, 15 STRUCTURES | | Descriptor: | PEPTIDE GVKESLV, SYNTROPHIN | | Authors: | Schultz, J, Hoffmueller, U, Ashurst, J, Krause, G, Schmieder, P, Macias, M, Schneider-Mergener, J, Oschkinat, H. | | Deposit date: | 1997-12-10 | | Release date: | 1998-12-30 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels.
Nat.Struct.Biol., 5, 1998
|
|
1R2N
 
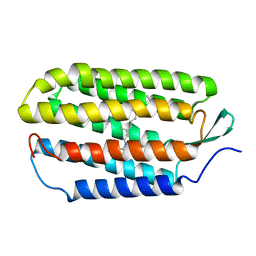 | | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, terLaak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1R84
 
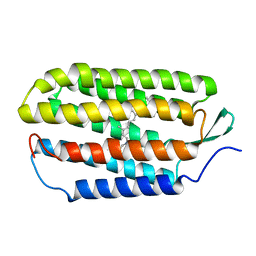 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2RQK
 
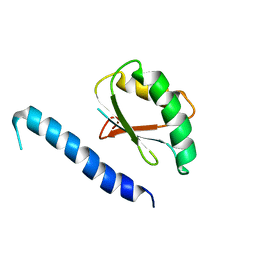 | | NMR Solution Structure of Mesoderm Development (MESD) - closed conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
2RQM
 
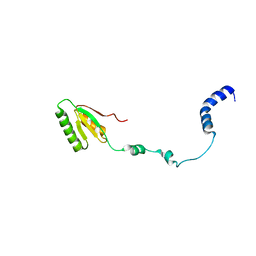 | | NMR Solution Structure of Mesoderm Development (MESD) - open conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-14 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
1OQA
 
 | | Solution structure of the BRCT-c domain from human BRCA1 | | Descriptor: | Breast cancer type 1 susceptibility protein | | Authors: | Gaiser, O.J, Ball, L.J, Schmieder, P, Leitner, D, Strauss, H, Wahl, M, Kuhne, R, Oschkinat, H, Heinemann, U. | | Deposit date: | 2003-03-07 | | Release date: | 2004-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and association behavior of the C-terminal BRCT domain from the breast cancer-associated protein BRCA1.
Biochemistry, 43, 2004
|
|
