7TJK
 
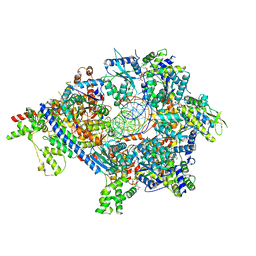 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJJ
 
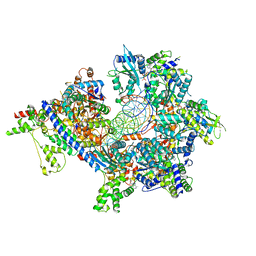 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJH
 
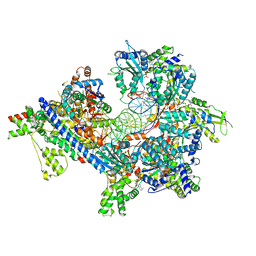 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJI
 
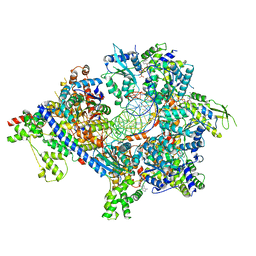 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJF
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7JK4
 
 | | Structure of Drosophila ORC bound to AT-rich DNA and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein, DNA (34-MER), ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JGS
 
 | |
7JK6
 
 | | Structure of Drosophila ORC in the active conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK2
 
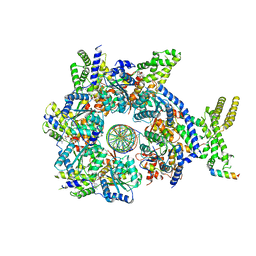 | |
7JGR
 
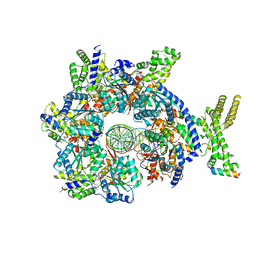 | | Structure of Drosophila ORC bound to DNA (84 bp) and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AT22044p1, Cell division control protein, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK3
 
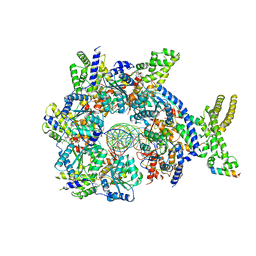 | | Structure of Drosophila ORC bound to GC-rich DNA and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein, DNA (33-MER), ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK5
 
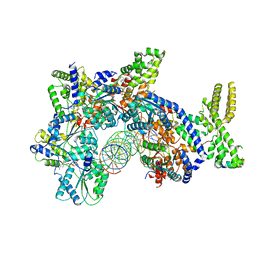 | | Structure of Drosophila ORC bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (32-MER), MAGNESIUM ION, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
2K8V
 
 | | Solution structure of Oxidised ERp18 | | Descriptor: | Thioredoxin domain-containing protein 12 | | Authors: | Rowe, M.L, Alanen, H.I, Ruddock, L.W, Kelly, G, Schmidt, J.M, Williamson, R.A, Howard, M.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-06-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase .
Biochemistry, 48, 2009
|
|
3GGX
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-4-{[(2S)-3,3-dimethyl-2-{3-[(6-methylpyridin-2-yl)methyl]-2-oxo-2,3-dihydro-1H-imidazol-1-yl}butanoyl]amino}-3-hydroxy-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3GGV
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-3-hydroxy-4-{[(2S)-2-(3-{[6-(1-hydroxy-1-methylethyl)pyridin-2-yl]methyl}-2-oxo-2,3-dihydro-1H-imidazol-1-yl)-3,3-dimethylbutanoyl]amino}-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3GGA
 
 | | HIV Protease inhibitors with pseudo-symmetric cores | | Descriptor: | V-1 protease, methyl [(1S,4S,5S,7S,10S)-4-benzyl-1,10-di-tert-butyl-5-hydroxy-2,9,12-trioxo-7-(4-pyridin-2-ylbenzyl)-13-oxa-3,8,11-triazatetradec-1-yl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3S85
 
 | | Discovery of New HIV Protease Inhibitors with Potential for Convenient Dosing and Reduced Side Effects: A-790742 and A-792611. | | Descriptor: | Protease/reverse transcriptase, methyl N-[(2S)-1-[[(2S,3S,5S)-5-[[(2S)-2-(methoxycarbonylamino)-3,3-dimethyl-butanoyl]amino]-3-oxidanyl-6-phenyl-1-(4-pyridin-3-ylphenyl)hexan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeGoey, D.A, Flosi, W.J, Grampovnik, D.J, Flentge, C.A. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus protease inhibitors (A-792611 and A-790742) with potential for convenient dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
1EAL
 
 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
1YGW
 
 | |
