4RH7
 
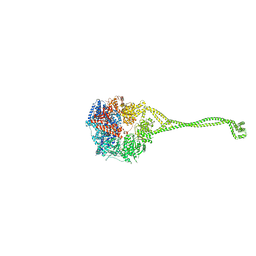 | | Crystal structure of human cytoplasmic dynein 2 motor domain in complex with ADP.Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, ... | | Authors: | Schmidt, H, Zalyte, R, Urnavicius, L, Carter, A.P. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of human cytoplasmic dynein-2 primed for its power stroke.
Nature, 518, 2015
|
|
2XCI
 
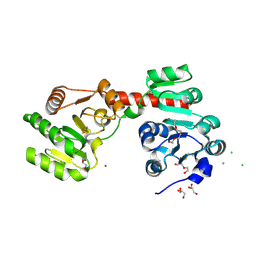 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XCU
 
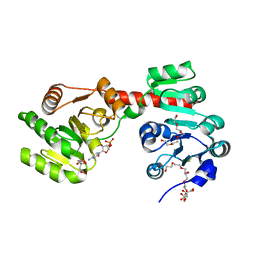 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2OI3
 
 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Tyrosine-protein kinase HCK, artificial peptide PD1 | | Authors: | Schmidt, H, Stoldt, M, Hoffmann, S, Tran, T, Willbold, D. | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand
Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
2OJ2
 
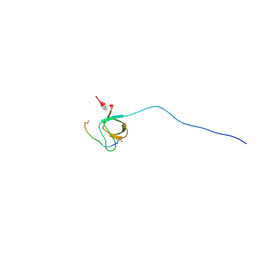 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Hematopoetic Cell Kinase, SH3 domain, artificial peptide PD1 | | Authors: | Schmidt, H, Hoffmann, S, Tran, T, Stoldt, M, Stangler, T, Wiesehan, K, Willbold, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
2Y6P
 
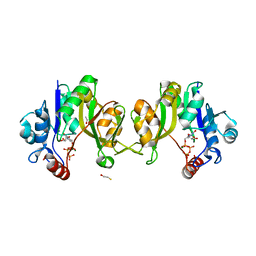 | | Evidence for a Two-Metal-Ion-Mechanism in the Kdo- Cytidylyltransferase KdsB | | Descriptor: | 3-DEOXY-MANNO-OCTULOSONATE CYTIDYLYLTRANSFERASE, BETA-MERCAPTOETHANOL, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Schmidt, H, Mesters, J.R, Mamat, U, Hilgenfeld, R. | | Deposit date: | 2011-01-25 | | Release date: | 2011-08-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for a Two-Metal-Ion Mechanism in the Cytidyltransferase Kdsb, an Enzyme Involved in Lipopolysaccharide Biosynthesis.
Plos One, 6, 2011
|
|
4AKG
 
 | | Dynein Motor Domain - ATP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKH
 
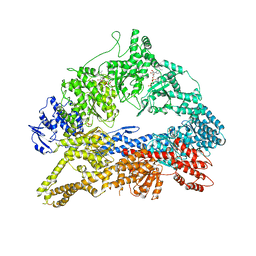 | | Dynein Motor Domain - AMPPNP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, DYNEIN HEAVY CHAIN CYTOPLASMIC, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AI6
 
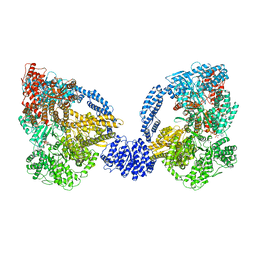 | | Dynein Motor Domain - ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-14 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKI
 
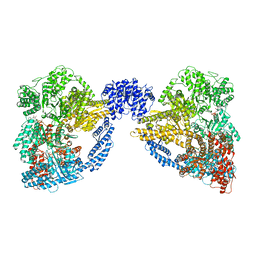 | | Dynein Motor Domain - LuAc derivative | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, DYNEIN HEAVY CHAIN CYTOPLASMIC, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
2KV7
 
 | | NMR solution structure of a soluble PrgI mutant from Salmonella Typhimurium | | Descriptor: | Protein prgI | | Authors: | Schmidt, H, Poyraz, O, Seidel, K, Delissen, F, Ader, C, Tenenboim, H, Goosmann, C, Laube, B, Thuenemann, A.F, Zychlinski, A, Baldus, M, Lange, A, Griesinger, C, Kolbe, M. | | Deposit date: | 2010-03-09 | | Release date: | 2010-06-16 | | Last modified: | 2019-12-18 | | Method: | SOLUTION NMR | | Cite: | Protein refolding is required for assembly of the type three secretion needle.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CJY
 
 | | HUMAN CYTOSOLIC PHOSPHOLIPASE A2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, PROTEIN (CYTOSOLIC PHOSPHOLIPASE A2) | | Authors: | Dessen, A, Tang, J, Schmidt, H, Stahl, M, Clark, J.D, Seehra, J, Somers, W.S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
2X9C
 
 | | Crystal structure of a soluble PrgI mutant from Salmonella Typhimurium | | Descriptor: | PROTEIN PRGI | | Authors: | Poyraz, O, Schmidt, H, Seidel, K, Delissen, F, Ader, C, Tenenboim, H, Goosmann, C, Laube, B, Thuenemann, A.F, Zychlinsky, A, Baldus, M, Lange, A, Griesinger, C, Kolbe, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-06-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein Refolding is Required for Assembly of the Type Three Secretion Needle
Nat.Struct.Mol.Biol., 17, 2010
|
|
1R26
 
 | | Crystal structure of thioredoxin from Trypanosoma brucei brucei | | Descriptor: | Thioredoxin | | Authors: | Friemann, R, Schmidt, H, Ramaswamy, S, Forstner, M, Krauth-Siegel, R.L, Eklund, H. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of thioredoxin from Trypanosoma brucei brucei
FEBS Lett., 554, 2003
|
|
6HYD
 
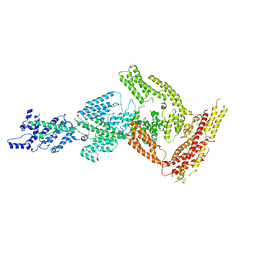 | | Rea1 Wild type ADP state (tail part) | | Descriptor: | Midasin,Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-19 | | Release date: | 2018-12-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
6HYP
 
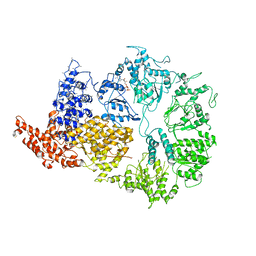 | | Rea1 Wild type ADP state (AAA+ ring part) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
6I26
 
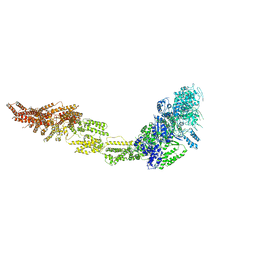 | | Rea1 Wild type AMPPNP state | | Descriptor: | Midasin,Midasin,Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
6I27
 
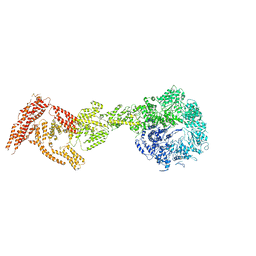 | | Rea1 AAA2L-H2alpha deletion mutant in AMPPNP State | | Descriptor: | Midasin,Midasin,Midasin,Midasin,Midasin,Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
1DB3
 
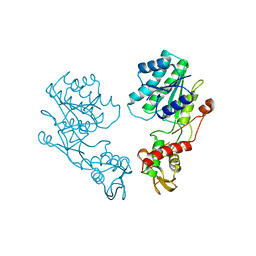 | | E.COLI GDP-MANNOSE 4,6-DEHYDRATASE | | Descriptor: | GDP-MANNOSE 4,6-DEHYDRATASE | | Authors: | Somoza, J.R, Menon, S, Somers, W.S, Sullivan, F.X. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose.
Structure Fold.Des., 8, 2000
|
|
3JZ7
 
 | |
5K8Y
 
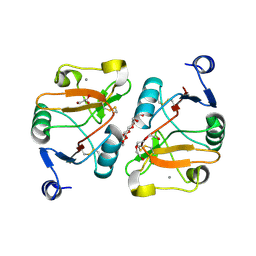 | | Structure of the Mus musclus Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
5M62
 
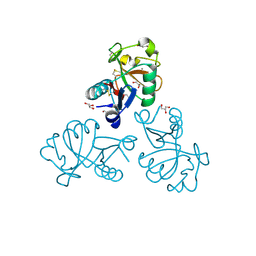 | | Structure of the Mus musclus Langerin carbohydrate recognition domain in complex with glucose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-10-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
2LEV
 
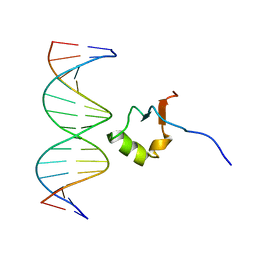 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
2K7Y
 
 | |
