2W03
 
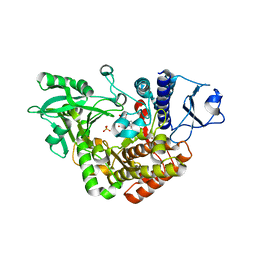 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with adenosine, sulfate and citrate from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE, CITRIC ACID, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W04
 
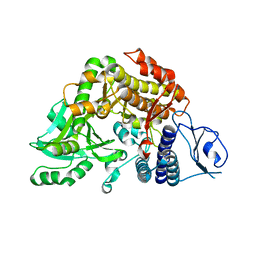 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W02
 
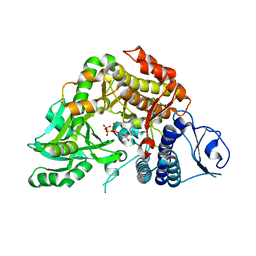 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with ATP from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
Nat. Chem. Biol., 5, 2009
|
|
2X3J
 
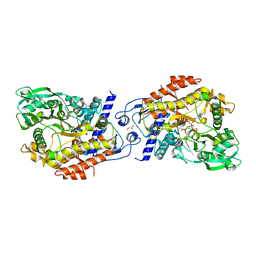 | | CO-COMPLEX STRUCTURE OF ACHROMOBACTIN SYNTHETASE PROTEIN D (ACSD) WITH ATP AND N-CITRYL-ETHYLENEDIAMINE FROM PECTOBACTERIUM CHRYSANTHEMI | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, ACSD, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schmelz, S, Challis, G.L, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Acyl Acceptor Specificity in the Achromobactin Biosynthetic Enzyme Acsd.
J.Mol.Biol., 412, 2011
|
|
2X0P
 
 | |
2X3K
 
 | |
2X0Q
 
 | |
5FZP
 
 | | Structure of the dispase autolysis inducing protein from Streptomyces mobaraensis | | Descriptor: | CALCIUM ION, DISPASE AUTOLYSIS-INDUCING PROTEIN, GLYCEROL | | Authors: | Schmelz, S, Fiebig, D, Beck, J, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2016-03-15 | | Release date: | 2016-08-10 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
J.Biol.Chem., 291, 2016
|
|
7NBW
 
 | |
3FFE
 
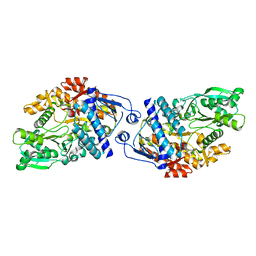 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
6GMG
 
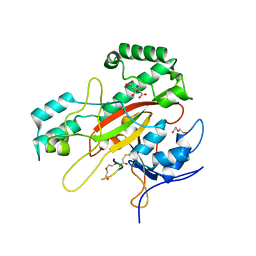 | | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, ... | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase.
FEBS J., 285, 2018
|
|
6I0I
 
 | |
6YA1
 
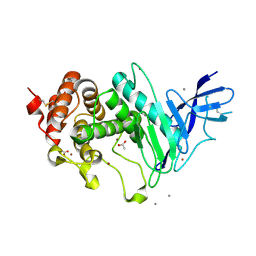 | | Zinc metalloprotease ProA | | Descriptor: | ACETATE ION, CALCIUM ION, ZINC ION, ... | | Authors: | Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2020-03-11 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Zinc metalloprotease ProA of Legionella pneumophila increases alveolar septal thickness in human lung tissue explants by collagen IV degradation.
Cell.Microbiol., 23, 2021
|
|
6YIZ
 
 | |
6YZE
 
 | | Zinc metalloprotease ProA from native source | | Descriptor: | ZINC ION, Zinc metalloproteinase | | Authors: | Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2020-05-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Zinc metalloprotease ProA of Legionella pneumophila increases alveolar septal thickness in human lung tissue explants by collagen IV degradation.
Cell.Microbiol., 23, 2021
|
|
5OJY
 
 | | Co-complex structure of regulator protein 2 (PamR2) with pamamycin 607 from Streptomyces alboniger | | Descriptor: | CITRIC ACID, GLYCEROL, Pamamycin 607, ... | | Authors: | Schmelz, S, Rebets, Y, Luzhetskyy, A, Scrima, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Design, development and application of whole-cell based antibiotic-specific biosensor.
Metab. Eng., 47, 2018
|
|
5OJX
 
 | | Crystal structure of regulator protein 2 (PamR2) from the pamamycin biosynthetic gene cluster of Streptomyces alboniger | | Descriptor: | TetR family transcription regulator | | Authors: | Schmelz, S, Rebets, Y, Luzhetskyy, A, Scrima, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, development and application of whole-cell based antibiotic-specific biosensor.
Metab. Eng., 47, 2018
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
8BTJ
 
 | | Murine cytomegalovirus protein M35 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MALONATE ION, Protein M35 | | Authors: | Schmelz, S, Van den Heuvel, J, Blankenfeldt, W. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Cytomegalovirus M35 Protein Directly Binds to the Interferon-beta Enhancer and Modulates Transcription of Ifnb1 and Other IRF3-Driven Genes.
J.Virol., 97, 2023
|
|
5NTB
 
 | | Streptomyces papain inhibitor (SPI) | | Descriptor: | Papain inhibitor, SULFATE ION | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Illuminating structure and acyl donor sites of a physiological transglutaminase substrate from Streptomyces mobaraensis.
Protein Sci., 27, 2018
|
|
4ALZ
 
 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
4A1V
 
 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4A1X
 
 | | Co-Complex structure of NS3-4A protease with the inhibitory peptide CP5-46-A (Synchrotron data) | | Descriptor: | CHLORIDE ION, CP5-46-A PEPTIDE, NONSTRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4AOQ
 
 | | Cationic trypsin in complex with mutated Spinacia oleracea trypsin inhibitor III (SOTI-III) (F14A) | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
