2MTZ
 
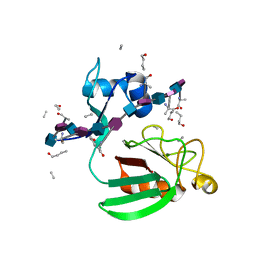 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
4XOK
 
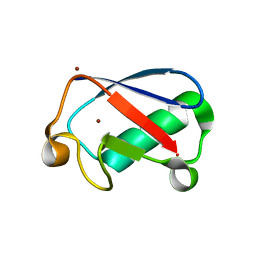 | | Observing the overall rocking motion of a protein in a crystal. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Ma, P, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6F3K
 
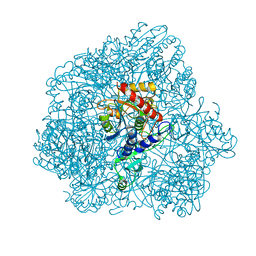 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6HWM
 
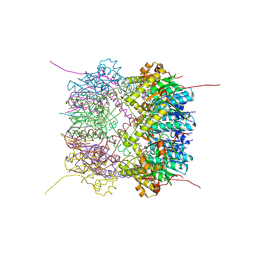 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
4XOF
 
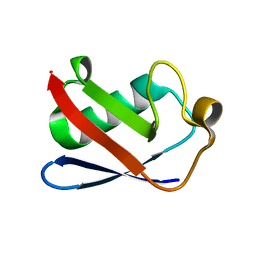 | |
4XOL
 
 | | Observing the overall rocking motion of a protein in a crystal - Cubic Ubiquitin crystals. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Peixiang, M, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
6HWN
 
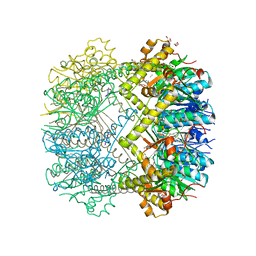 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
2K0Q
 
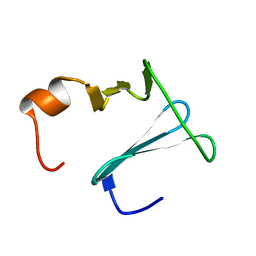 | | Solution structure of CopK, a periplasmic protein involved in copper resistance in Cupriavidus metallidurans CH34 | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Bersch, B, Favier, A, Schanda, P, Coves, J, van Aelst, S, Vallaeys, T, Wattiez, R, Mergeay, M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Molecular structure and metal-binding properties of the periplasmic CopK protein expressed in Cupriavidus metallidurans CH34 during copper challenge.
J.Mol.Biol., 380, 2008
|
|
2L3Z
 
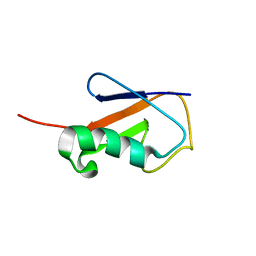 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
8CQR
 
 | |
2KM0
 
 | | Cu(I)-bound CopK | | Descriptor: | COPPER (I) ION, Copper resistance protein K | | Authors: | Bersch, B. | | Deposit date: | 2009-07-15 | | Release date: | 2010-03-16 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | CopK from Cupriavidus metallidurans CH34 Binds Cu(I) in a Tetrathioether Site: Characterization by X-ray Absorption and NMR Spectroscopy
J.Am.Chem.Soc., 2010
|
|
2LXS
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2LXT
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
